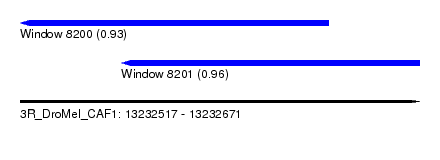
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,232,517 – 13,232,671 |
| Length | 154 |
| Max. P | 0.964342 |

| Location | 13,232,517 – 13,232,636 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -23.79 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.05 |
| Structure conservation index | 0.69 |
| SVM decision value | 1.21 |
| SVM RNA-class probability | 0.931015 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13232517 119 - 27905053 AUUGAACUGGUUCUGAAUACAGCUGUUAAAAUUCAUUAACGUUUAAUAGUGUUCAGAUAAAUAUGAAUACAGUUCAACUCUUUUUUGAAUGUGAUGGAGUUCAGC-AUCCAUUACCUAUG .((((((((..((((((((((((.(((((......)))))))).....)))))))))............)))))))).............((((((((.......-.))))))))..... ( -29.84) >DroSec_CAF1 261211 104 - 1 AUAGAACUGGUUCUGAAUUCAGCUGUUCAAUUUCAUUAACGUUUAAUAGUGUUCAGAU-------AAUACAGUUCAACUCUUUUUUAUAUG--------UUCCGC-AGCCAUUACCUAUG ...((((((..(((((((...((((((.(((.........))).))))))))))))).-------....))))))................--------......-.............. ( -18.30) >DroSim_CAF1 246690 104 - 1 AUAGAACUGGUUCUGAAUACAGCUGUUAAAUUUCAUUAACGUUUAAUAGUGUUCAAAA-------AAUACAGUUCAACUCUUUUUUAUAUG--------UUCCGC-AGCCAUUACCUAUG ...((((((.((.(((((...((((((((((.........))))))))))))))).))-------....))))))................--------......-.............. ( -17.70) >DroEre_CAF1 250899 105 - 1 ---GAACUGGUUCUGAUUACAGCUGUUAAACUCAAUGAACAUUUAAUAGUGUUCAGAUAA------AUACAGUUCAACCCGUUUUUGUAUGGGA------UCAGCUAUUAAUUACCUAUG ---((((((..(((((.....(((((((((...........)))))))))..)))))...------...))))))..(((((......))))).------.................... ( -24.80) >DroYak_CAF1 261728 113 - 1 GAUGAACUGGUUCUGAAUACAGCUGUUAAAUUCCAUGAACAUUUAAUAGUGUUCAGAUAAAUAUAAAUACAGUUCAAC-UGUUUUUGUAUGUAA------UCGCUUUUUCAUUGCCUAUG ........(((..((((...(((............((((((((....))))))))(((..(((((((.((((.....)-))).)))))))...)------)))))..))))..))).... ( -28.30) >consensus AUAGAACUGGUUCUGAAUACAGCUGUUAAAUUUCAUUAACGUUUAAUAGUGUUCAGAUAA_____AAUACAGUUCAACUCUUUUUUGUAUG__A_____UUCAGC_AUCCAUUACCUAUG ...((((((..(((((((...((((((((((.........)))))))))))))))))............))))))............................................. (-16.40 = -17.32 + 0.92)

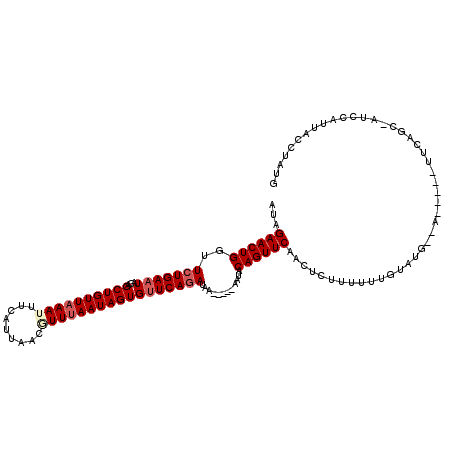
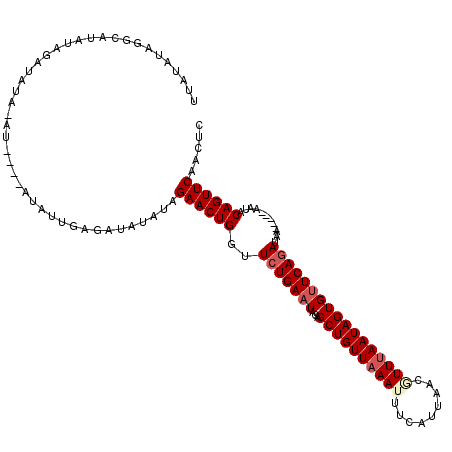
| Location | 13,232,556 – 13,232,671 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 81.15 |
| Mean single sequence MFE | -23.42 |
| Consensus MFE | -16.40 |
| Energy contribution | -17.32 |
| Covariance contribution | 0.92 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.57 |
| SVM RNA-class probability | 0.964342 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13232556 115 - 27905053 UUAUAUAGGCAUAUAAAUACAUAU----AUAUU-AGGUAUAUUGAACUGGUUCUGAAUACAGCUGUUAAAAUUCAUUAACGUUUAAUAGUGUUCAGAUAAAUAUGAAUACAGUUCAACUC .(((((....(((((........)----)))).-..)))))((((((((..((((((((((((.(((((......)))))))).....)))))))))............))))))))... ( -22.04) >DroSec_CAF1 261242 113 - 1 UUAUAUAGGCAUAUAGAUAUACAUAUGUAUAUUGAGUUAUAUAGAACUGGUUCUGAAUUCAGCUGUUCAAUUUCAUUAACGUUUAAUAGUGUUCAGAU-------AAUACAGUUCAACUC .(((((((.(.....(((((((....)))))))..))))))))((((((..(((((((...((((((.(((.........))).))))))))))))).-------....))))))..... ( -25.40) >DroSim_CAF1 246721 113 - 1 UUAUAUAGGCAUAUAGAUAUACAUAUGUAUAUUGAGAUAUAUAGAACUGGUUCUGAAUACAGCUGUUAAAUUUCAUUAACGUUUAAUAGUGUUCAAAA-------AAUACAGUUCAACUC .((((((........(((((((....)))))))....))))))((((((.((.(((((...((((((((((.........))))))))))))))).))-------....))))))..... ( -24.90) >DroEre_CAF1 250933 102 - 1 UUUUAUAAGCAUUUAGAUAUA--------UAUUGAUAUA----GAACUGGUUCUGAUUACAGCUGUUAAACUCAAUGAACAUUUAAUAGUGUUCAGAUAA------AUACAGUUCAACCC .................((((--------(....)))))----((((((..(((((.....(((((((((...........)))))))))..)))))...------...))))))..... ( -19.00) >DroYak_CAF1 261762 111 - 1 UUAUAUAGGCAUAUAGAUAUA--------UAUUGAUAUAUGAUGAACUGGUUCUGAAUACAGCUGUUAAAUUCCAUGAACAUUUAAUAGUGUUCAGAUAAAUAUAAAUACAGUUCAAC-U (((((((...(((((....))--------)))...)))))))(((((((..(((((((...((((((((((.........)))))))))))))))))............)))))))..-. ( -25.74) >consensus UUAUAUAGGCAUAUAGAUAUA_AU____AUAUUGAGAUAUAUAGAACUGGUUCUGAAUACAGCUGUUAAAUUUCAUUAACGUUUAAUAGUGUUCAGAUAA_____AAUACAGUUCAACUC ...........................................((((((..(((((((...((((((((((.........)))))))))))))))))............))))))..... (-16.40 = -17.32 + 0.92)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:23 2006