| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,884,308 – 1,884,413 |
| Length | 105 |
| Max. P | 0.696160 |

| Location | 1,884,308 – 1,884,413 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 78.95 |
| Mean single sequence MFE | -24.95 |
| Consensus MFE | -15.97 |
| Energy contribution | -15.92 |
| Covariance contribution | -0.05 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.696160 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1884308 105 - 27905053 CUUCCAGAUAAAAAGGUGAUUGACGAAGCGAAAGCAAAAGUCAUGGUCGAA-CUUCCACUUAGCA-AUGCAGUUGUCGUUCACA-G-AAAAGGACCAACGACGAUGUCG .....................((((..((....))....(((.((((((((-(..(.(((((...-.)).))).)..)))).(.-.-....))))))..)))..)))). ( -22.90) >DroVir_CAF1 43777 105 - 1 CUGCAAGAUAAAAAGGUGAUUGACGAACCGAAAGCAAAAGUCAUGGCCAAAGCUUUGGCUCAGCAAAUGCAUUUGUCGUUUGCCCGUAAAACUAGUC---AAAAUGUG- ..(((.........(((.((.(((....(....).....))))).))).....(((((((..((((((((....).)))))))..(.....).))))---))).))).- ( -21.20) >DroPse_CAF1 32792 95 - 1 GUCCCAGAUAAAAAGGUGAUUGACGAAGCGAAAGCAAAAGUCAUGGCCAAA-AUUCCACUUAGCA-AUGCAGUUGUCGUUGACU-G-AGAAAGAAAGGG---------- ..(((.........(((.((.(((...((....))....))))).)))...-......(((((((-((((....).))))).))-)-)).......)))---------- ( -26.80) >DroEre_CAF1 43073 101 - 1 CUUCCAGAUAAAAAGGUGAUUGACGAAGCGAAAGCAAAAGUCAUGGUCGAA-CUUCCACUUAGCA-AUGCAGUUGUCGUUCGCA---AAAAGGACGAAC---GAUGUGG ................((((((((...((....))....))))..))))..-...((((...((.-..))....(((((((((.---.....).)))))---))))))) ( -24.00) >DroAna_CAF1 31629 102 - 1 CUUUCAGAUAAAAAGGUGAUUGACGAAGCGAAAGCAAAAGUCAUGGCCGAA-AUUCCACUUAGCA-AUGCAGUUGUCGUUUUCG-G-AGAAGGACGAAAGUCC---UGC .((((.........(((.((.(((...((....))....))))).)))(((-(..(.(((((...-.)).))).)....)))).-)-)))(((((....))))---).. ( -28.00) >DroPer_CAF1 33112 95 - 1 GUCCCAGAUAAAAAGGUGAUUGACGAAGCGAAAGCAAAAGUCAUGGCCAAA-AUUCCACUUAGCA-AUGCAGUUGUCGUUGACU-G-AGAAAGAAAGGG---------- ..(((.........(((.((.(((...((....))....))))).)))...-......(((((((-((((....).))))).))-)-)).......)))---------- ( -26.80) >consensus CUUCCAGAUAAAAAGGUGAUUGACGAAGCGAAAGCAAAAGUCAUGGCCAAA_AUUCCACUUAGCA_AUGCAGUUGUCGUUCACA_G_AAAAGGACAAAC_______UG_ ......(((((...(((.((.(((...((....))....))))).)))..............((....))..)))))................................ (-15.97 = -15.92 + -0.05)

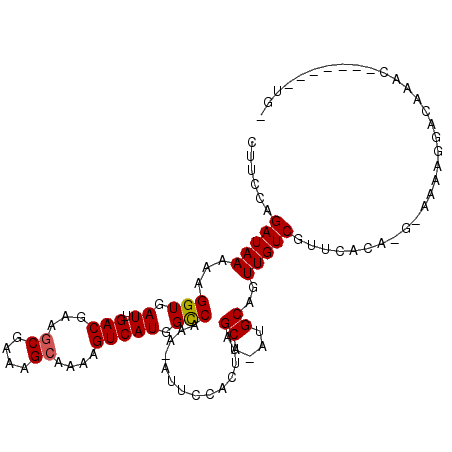
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:42 2006