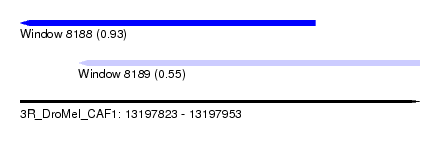
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,197,823 – 13,197,953 |
| Length | 130 |
| Max. P | 0.926824 |

| Location | 13,197,823 – 13,197,919 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 80.97 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -19.41 |
| Energy contribution | -19.97 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.18 |
| SVM RNA-class probability | 0.926824 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
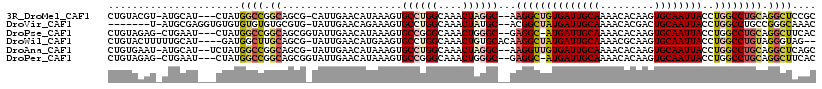
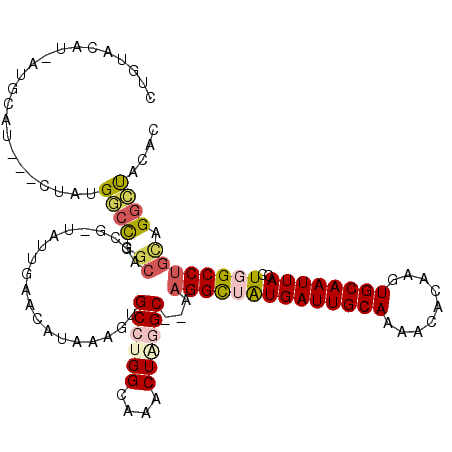
>3R_DroMel_CAF1 13197823 96 - 27905053 AACAUAAAGUGCCUGGCAAACUAGGC--AAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCCGCA------UAUACACAAACACUCAGG--- .........(((((((....))))))--)((((((((((((((.........)))))))))..)))))((.(....))).------..................--- ( -27.10) >DroVir_CAF1 227451 96 - 1 AACAGAAAGUGCCUGGCAAACUAUGC--ACGGCUAUGAUUGCAAAACACGACUGCAAUUACCUGGCCUGCCGGGCAAACUCAGU------CGCACACUCUCACA--- ....((...(((((((((........--..(((((((((((((.........))))))))..))))))))))))))...))...------..............--- ( -29.70) >DroSec_CAF1 225673 96 - 1 AACAUAAAGUGCCUGGCAAACUAGGC--AAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCCGCA------UAUACACAAACACCCAGG--- .........(((((((....))))))--)((((((((((((((.........)))))))))..)))))((.(....))).------..................--- ( -27.10) >DroEre_CAF1 216286 96 - 1 AACAUAAAGUGCCUGGCAAACUAGGC--AAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCGGCA------CAUACACAAACACGCAGG--- ........(((((.(((.......((--(.(((((((((((((.........)))))))))..)))))))..))).))))------).................--- ( -31.90) >DroWil_CAF1 212494 95 - 1 AACAUGAAGUGCCUGGCAAACUGUGCACAAGGCUAUGAUUGCAAAACGCAAGUGCAAUUACCUGGCCUGUAGGGUAG------------ACACAAAUACUCAUGCAC ........((((.(((....))).)))).(((((((((((((...((....)))))))))..))))))((((((((.------------.......))))).))).. ( -28.40) >DroAna_CAF1 211789 102 - 1 AACAUAAAGUGCCUGGCAAACUAGGC--AAGGUUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCAGCACGCUCACACACUCAACCCCGCACG--- .........(((((((....))))))--).(((((((((((((.........)))))))))((((((....))).)))...............)))).......--- ( -29.30) >consensus AACAUAAAGUGCCUGGCAAACUAGGC__AAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCAGCA_______AUACACAAACACUCAGG___ ..........((((((....))))))...((((((((((((((.........)))))))))..)))))....................................... (-19.41 = -19.97 + 0.56)

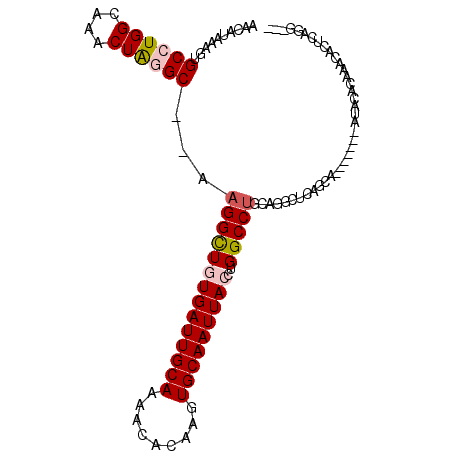
| Location | 13,197,842 – 13,197,953 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | reverse |
| Mean pairwise identity | 79.79 |
| Mean single sequence MFE | -35.97 |
| Consensus MFE | -21.43 |
| Energy contribution | -22.13 |
| Covariance contribution | 0.70 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.60 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.554388 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13197842 111 - 27905053 CUGUACGU-AUGCAU---CUAUGGCCGGCAGCG-CAUUGAACAUAAAGUGCCUGGCAAACUAGGC--AAGGCUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCCGC .((((...-.)))).---....(((((.(((.(-((((........)))))))).).......((--(.(((((((((((((.........)))))))))..))))))).)))).... ( -36.30) >DroVir_CAF1 227470 107 - 1 -------U-AUGCGAGGUGUGUGUGUGUGCGUG-UAUUGAACAGAAAGUGCCUGGCAAACUAUGC--ACGGCUAUGAUUGCAAAACACGACUGCAAUUACCUGGCCUGCCGGGCAAAC -------.-.(((..(((((((((((.(((..(-((((........)))))...))).)).))))--))(((((((((((((.........))))))))..))))).)))..)))... ( -34.10) >DroPse_CAF1 217295 111 - 1 CUGUAGAG-CUGAAU---CUAUGGCCGGCAGCGGUAUUGAACAUAAAGUGCCGGGCAAACUGGGC--GAGGC-AUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUUCAC ..(((((.-.....)---))))((((.((..(((((((........)))))))........((((--.(((.-.((((((((.........))))))))))).)))))).)))).... ( -38.60) >DroWil_CAF1 212512 111 - 1 CUGUACUUUUUGCAU----GAUGGCUUGCAGCG-UAUUGAACAUGAAGUGCCUGGCAAACUGUGCACAAGGCUAUGAUUGCAAAACGCAAGUGCAAUUACCUGGCCUGUAGGGUAG-- ((.(((....(((((----...((.((((((.(-((((........))))))).)))).)))))))..(((((((((((((...((....)))))))))..))))))))).))...-- ( -33.30) >DroAna_CAF1 211814 112 - 1 CUGUGAAU-AUGCAU--UCUAUGGCCGGCAGCG-UAUUGAACAUAAAGUGCCUGGCAAACUAGGC--AAGGUUGUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUCAGC (((.(...-.((((.--.....((((((.....-.....(((......(((((((....))))))--)..)))(((((((((.........)))))))))))))))))))..).))). ( -34.90) >DroPer_CAF1 283962 111 - 1 CUGUAGAG-CUGAAU---CUAUGGCCGGCAGCGGUAUUGAACAUAAAGUGCCGGGCAAACUGGGC--GAGGC-AUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUUCAC ..(((((.-.....)---))))((((.((..(((((((........)))))))........((((--.(((.-.((((((((.........))))))))))).)))))).)))).... ( -38.60) >consensus CUGUACAU_AUGCAU___CUAUGGCCGGCAGCG_UAUUGAACAUAAAGUGCCUGGCAAACUAGGC__AAGGCUAUGAUUGCAAAACACAAGUGCAAUUACCUGGCCUGCAGGCUACAC ......................((((.((....................((((((....))))))...((((((((((((((.........))))))))..)))))))).)))).... (-21.43 = -22.13 + 0.70)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:11 2006