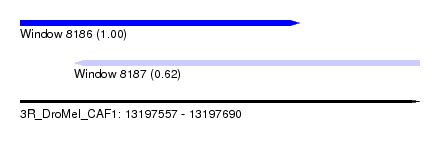
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,197,557 – 13,197,690 |
| Length | 133 |
| Max. P | 0.998801 |

| Location | 13,197,557 – 13,197,650 |
|---|---|
| Length | 93 |
| Sequences | 4 |
| Columns | 93 |
| Reading direction | forward |
| Mean pairwise identity | 79.14 |
| Mean single sequence MFE | -24.45 |
| Consensus MFE | -17.52 |
| Energy contribution | -18.77 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.04 |
| Mean z-score | -2.93 |
| Structure conservation index | 0.72 |
| SVM decision value | 3.23 |
| SVM RNA-class probability | 0.998801 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
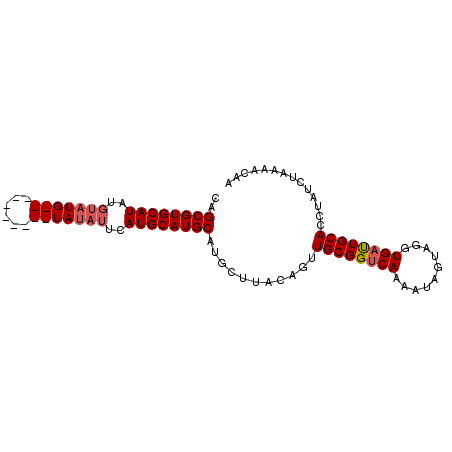
>3R_DroMel_CAF1 13197557 93 + 27905053 CAGCGUGCAUAUGUAUGUAUAUGUAUGUAUAUUCAUGCAUGCAUGCUUACAGUUGCGGUCAAAAUAGUAGGUGAUUGCACGUAUCUAAAACAA ..(((((((.((((((((((..(((....)))..))))))))))((((((.(((........))).))))))...)))))))........... ( -27.40) >DroSec_CAF1 225421 84 + 1 CAGCGUGCAUAUGUAUGU--------GUAUAUUCAUGCAUGCAAGCUUACAGUUGCGGUCAAAAUAGUAGGUGAUUGCACCUAACUAAAAA-G ..((((((((..((((..--------..))))..))))))))..((........))........(((((((((....))))).))))....-. ( -24.10) >DroSim_CAF1 211903 83 + 1 CAGCGUGCAUAUGUAUG----------UAUAUCCAUGCAUGCAUGCUUACAGUUGCGGUCAAAAUAGUAGGUGAUUGCACCUAUAUAAAACAA .((((((((..((((((----------......))))))))))))))...((.((((((((..........)))))))).))........... ( -25.00) >DroYak_CAF1 225250 73 + 1 CAGCGUGCAUA------------------UAUUCAUGCAUGCCUGCUUACACUUGCGGCCAAAAUAGUAGGUGCCUGCACUUU--CGAAACAG ..((((((((.------------------.....)))))))).(((...(((((((..........)))))))...)))....--........ ( -21.30) >consensus CAGCGUGCAUAUGUAUG__________UAUAUUCAUGCAUGCAUGCUUACAGUUGCGGUCAAAAUAGUAGGUGAUUGCACCUAUCUAAAACAA ..((((((((..(((((((......)))))))..))))))))...........((((((((..........)))))))).............. (-17.52 = -18.77 + 1.25)


| Location | 13,197,575 – 13,197,690 |
|---|---|
| Length | 115 |
| Sequences | 5 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 71.99 |
| Mean single sequence MFE | -27.56 |
| Consensus MFE | -6.20 |
| Energy contribution | -6.96 |
| Covariance contribution | 0.76 |
| Combinations/Pair | 1.30 |
| Mean z-score | -1.67 |
| Structure conservation index | 0.22 |
| SVM decision value | 0.18 |
| SVM RNA-class probability | 0.620346 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13197575 115 - 27905053 AUUGGUUGGUGCAUUGUAAACGCCUUUAAAGGUGGUUUCUUUGUUUUAGAUACGUGCAAUCACCUACUAUUUUGACCGCAACUGUAAGCAUGCAUGCAUGAAUAUACAUACAUAU ..((((.((((.((((((..(((((....)))))((.(((.......))).)).)))))))))).)))).............((((..((((....))))....))))....... ( -27.10) >DroSec_CAF1 225439 106 - 1 AUUUUUUGGUGCUUUGUAAACCCCUUUAAAGUUGGUUUCUC-UUUUUAGUUAGGUGCAAUCACCUACUAUUUUGACCGCAACUGUAAGCUUGCAUGCAUGAAUAUAC-------- .....((.((((..(((((....(((...(((((...((..-....((((.(((((....)))))))))....))...)))))..))).))))).)))).)).....-------- ( -22.00) >DroSim_CAF1 211920 106 - 1 AUUUUCUGUUGCAUUGUAAGCUCCUUUAAAUGUGGUUUCCUUGUUUUAUAUAGGUGCAAUCACCUACUAUUUUGACCGCAACUGUAAGCAUGCAUGCAUGGAUAUA--------- ....(((((.((((.(((.(((........(((((((..........(((((((((....)))))).)))...)))))))......))).))))))))))))....--------- ( -24.36) >DroEre_CAF1 216055 97 - 1 AUUAGCAUGCUCAGUGUAAGCG-CUCUAAAAGUGGUUUCUCUGUUUGGGCAAAGUGCAGGCAGCUACUCUUCUGGCCGCGACUGUAGGCGUGCAUACG----------------- ....(((((((((((....(((-((..((.(((((.....(((((((.((...)).)))))))))))).))..)).))).))))..))))))).....----------------- ( -31.50) >DroYak_CAF1 225261 101 - 1 AUGUGCAUGCUCAUUGUAAGCC-UUGUAAAAGUGCUUUCCCUGUUUCG--AAAGUGCAGGCACCUACUAUUUUGGCCGCAAGUGUAAGCAGGCAUGCAUGAAUA----------- ..(((((((((...(((..(((-((((....(..(((((........)--))))..).(((.............)))))))).))..)))))))))))).....----------- ( -32.82) >consensus AUUUGCUGGUGCAUUGUAAGCCCCUUUAAAAGUGGUUUCUCUGUUUUAGAUAAGUGCAAUCACCUACUAUUUUGACCGCAACUGUAAGCAUGCAUGCAUGAAUAUA_________ ..............((((.(((..((((...((((((.........(((....(((....)))...)))....)))))).....))))..))).))))................. ( -6.20 = -6.96 + 0.76)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:09 2006