
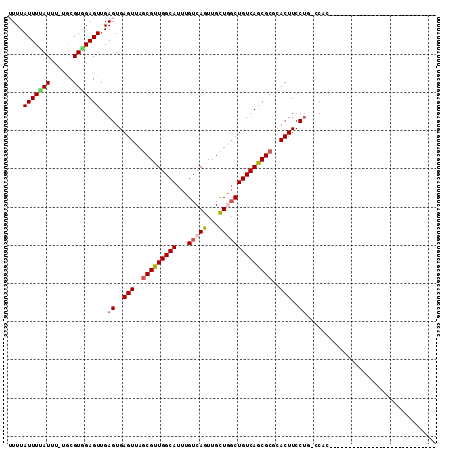
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,181,916 – 13,182,020 |
| Length | 104 |
| Max. P | 0.991084 |

| Location | 13,181,916 – 13,182,020 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | forward |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -28.87 |
| Consensus MFE | -19.64 |
| Energy contribution | -20.08 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.35 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.25 |
| SVM RNA-class probability | 0.991084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13181916 104 + 27905053 UUUUAUUUUAUUU-UGCGUGGAGUUGAGUGAGUUAGCGUUGGCAUUUGUCAGUUGCUAGCUGUCAGCGCUCACUUCCUG-CCACGCCCCUUUUCGCCACC------CCCUAC .............-.((((((....((((((....(((((((((.(((.(....).))).)))))))))))))))....-))))))..............------...... ( -30.40) >DroVir_CAF1 194502 83 + 1 UUUUAUUUCAUAU-UGCGUGGAGUUGAGUGAGUUAGCGCUGGCAUUUGUCAGUGGUUGCCUGUCAGCGCGCACUUCCUCCACUU---------------------------- .............-...((((((..(((((.....(((((((((...((........)).))))))))).))))).))))))..---------------------------- ( -29.50) >DroGri_CAF1 192368 83 + 1 UUUUAUUUAAUAU-UACGUGGAGUUGAGUGAGUUAACGUUGGCAUUUGUCAGUUGCUGGCUGUCAGCGCGCACUUCCUCCACUU---------------------------- .............-...((((((..(((((......((((((((...(((((...)))))))))))))..))))).))))))..---------------------------- ( -27.60) >DroSec_CAF1 209843 104 + 1 UUUUAUUUUAUUU-UGCGUGGAGUUGAGUGAGUUAGCGUUGGCAUUUGUCAGUUGCUAGCUGUCAGCGCUCACUUCCUG-CCACGCCCCCUUUCGCCAAC------CCCCAC .............-.((((((....((((((....(((((((((.(((.(....).))).)))))))))))))))....-))))))..............------...... ( -30.40) >DroWil_CAF1 195204 82 + 1 -UUUAUUUUAUUUCUGCGUAGAGUUGAGUGAGUUAGCGUUGGCAUUUGUCAGUUGCUGGCUGUCAGCGCGGACUUCC-G-CCACA--------------------------- -((.(((((((......))))))).))(..((((.(((((((((...(((((...)))))))))))))).))))..)-.-.....--------------------------- ( -24.40) >DroAna_CAF1 196274 105 + 1 UUUUAUUUCAUUU-UGCGUGGAGUUGAGUGAGUUAGCGUUGGCAUUUGUCAGUUGCUGGCUGUCAGCGCUUACUUCCUG-CC-----CCCAUCCAACCCGACCGCCCCCUUA .............-.((((((.((((((..(((.((((((((((...(((((...))))))))))))))).)))..)).-..-----......)))))))..)))....... ( -30.90) >consensus UUUUAUUUUAUUU_UGCGUGGAGUUGAGUGAGUUAGCGUUGGCAUUUGUCAGUUGCUGGCUGUCAGCGCGCACUUCCUG_CCAC____________________________ ....(((((((......)))))))..((..(((..(((((((((...(((((...))))))))))))))..)))..)).................................. (-19.64 = -20.08 + 0.45)


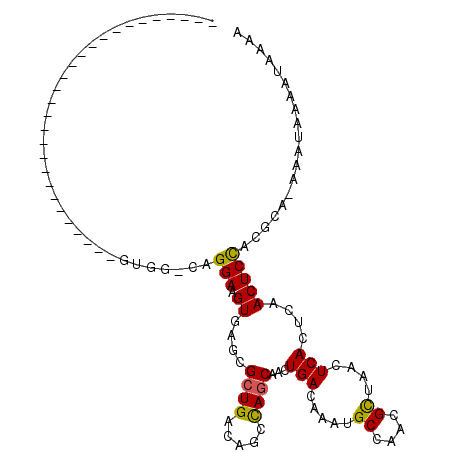
| Location | 13,181,916 – 13,182,020 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 74.48 |
| Mean single sequence MFE | -26.39 |
| Consensus MFE | -7.58 |
| Energy contribution | -7.25 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.29 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.783241 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13181916 104 - 27905053 GUAGGG------GGUGGCGAAAAGGGGCGUGG-CAGGAAGUGAGCGCUGACAGCUAGCAACUGACAAAUGCCAACGCUAACUCACUCAACUCCACGCA-AAAUAAAAUAAAA ((..((------(((..........(((((((-((....((.((.((((.....))))..)).))...)))).)))))..........)))))..)).-............. ( -28.55) >DroVir_CAF1 194502 83 - 1 ----------------------------AAGUGGAGGAAGUGCGCGCUGACAGGCAACCACUGACAAAUGCCAGCGCUAACUCACUCAACUCCACGCA-AUAUGAAAUAAAA ----------------------------..((((((..((((.((((((.((.(((.....)).)...)).)))))).....))))...))))))...-............. ( -25.00) >DroGri_CAF1 192368 83 - 1 ----------------------------AAGUGGAGGAAGUGCGCGCUGACAGCCAGCAACUGACAAAUGCCAACGUUAACUCACUCAACUCCACGUA-AUAUUAAAUAAAA ----------------------------..((((((..((((.(.((((.....))))...((((..........)))).).))))...))))))...-............. ( -20.20) >DroSec_CAF1 209843 104 - 1 GUGGGG------GUUGGCGAAAGGGGGCGUGG-CAGGAAGUGAGCGCUGACAGCUAGCAACUGACAAAUGCCAACGCUAACUCACUCAACUCCACGCA-AAAUAAAAUAAAA ((((((------(((((.(..((..(((((((-((....((.((.((((.....))))..)).))...)))).)))))..))..))))))))).))).-............. ( -37.00) >DroWil_CAF1 195204 82 - 1 ---------------------------UGUGG-C-GGAAGUCCGCGCUGACAGCCAGCAACUGACAAAUGCCAACGCUAACUCACUCAACUCUACGCAGAAAUAAAAUAAA- ---------------------------(((((-(-((....))))((((.....))))....................................)))).............- ( -16.30) >DroAna_CAF1 196274 105 - 1 UAAGGGGGCGGUCGGGUUGGAUGGG-----GG-CAGGAAGUAAGCGCUGACAGCCAGCAACUGACAAAUGCCAACGCUAACUCACUCAACUCCACGCA-AAAUGAAAUAAAA ...((((..(((.(((((((.((..-----((-((....((.((.((((.....))))..)).))...))))..))))))))))))...)))).....-............. ( -31.30) >consensus ____________________________GUGG_CAGGAAGUGAGCGCUGACAGCCAGCAACUGACAAAUGCCAACGCUAACUCACUCAACUCCACGCA_AAAUAAAAUAAAA ...................................(((.((....((((.....))))...(((.....((....))....)))....)))))................... ( -7.58 = -7.25 + -0.33)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:44:02 2006