
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,153,772 – 13,153,926 |
| Length | 154 |
| Max. P | 0.806959 |

| Location | 13,153,772 – 13,153,886 |
|---|---|
| Length | 114 |
| Sequences | 5 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 93.07 |
| Mean single sequence MFE | -28.92 |
| Consensus MFE | -24.80 |
| Energy contribution | -24.32 |
| Covariance contribution | -0.48 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.32 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.30 |
| SVM RNA-class probability | 0.677917 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
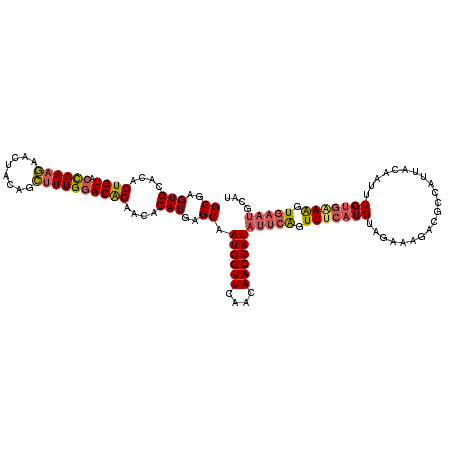
>3R_DroMel_CAF1 13153772 114 + 27905053 UCCGCCGUCAAAUUCCCCCUAAUGAGGGUCAGUCUGCGAGUGCACACUGUACCAAAGAACUACAGCUUUGGACAGAACACAUGAGCAAUCCUUCAACAAGGAUAUUCAGUUUCA ..(((((.((.(((..((((....))))..))).)))).)))....((((.((((((........))))))))))......((((..((((((....)))))).))))...... ( -29.30) >DroSec_CAF1 183141 114 + 1 UCCGCCGUCAAAUUUUUCCUAAUGAGGGUCAGUCUGCGAGUGCACACUGUACCAAAGAACUACAGCUUUGGACAGAACACAUGAGCCAUCCUUCAACAAGGAUAUUCGGUUUCA ...((((...(((...((((..((((((.......((..(((....((((.((((((........))))))))))....)))..))...))))))...)))).))))))).... ( -29.10) >DroSim_CAF1 168421 114 + 1 UCCGCCGUCAAAUUCCUCCUAAUGAGGGUCAGUCUGCGAGUGCACACUGUACCAAAUAUCUACAGAGUUGGACAGAACACAUGAGCCAUCCUUCAACAAGGAUAUUCAGUUUCA ......(((.((((((((.....)))(((((((.(((....))).)))).)))...........))))).))).(((.((.((((..((((((....)))))).))))))))). ( -29.00) >DroEre_CAF1 174266 114 + 1 UCCGCCGUCAAAUUCCCCCUAAUGAGGGUCAGCCUGCGAGUGCACACUGUCCUAAAGAACUACAGCUUUGGACAGAACACAUGAGCAAUCCUUCAACAAGGAUAUUCAGUUUCA ..(((((.((..((..((((....))))..))..)))).)))....((((((.((((........))))))))))......((((..((((((....)))))).))))...... ( -27.20) >DroYak_CAF1 181869 114 + 1 UCCGCCGUCAAAUGCCCCCUAAUGAGGGUCAGUCUGCGAGUGCACACUGUCCUAAAGAGCUACAGCUUUGCACGGAACACAUGGGCAAUCCUUCAACAAGGAUAUUCAGUUUCU ............(((((........(((.((((.(((....))).)))).)))..(((((....))))).............)))))((((((....))))))........... ( -30.00) >consensus UCCGCCGUCAAAUUCCCCCUAAUGAGGGUCAGUCUGCGAGUGCACACUGUACCAAAGAACUACAGCUUUGGACAGAACACAUGAGCAAUCCUUCAACAAGGAUAUUCAGUUUCA ..(((((.((.(((..((((....))))..))).)))).)))....((((.((((((........))))))))))......((((..((((((....)))))).))))...... (-24.80 = -24.32 + -0.48)



| Location | 13,153,807 – 13,153,926 |
|---|---|
| Length | 119 |
| Sequences | 5 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 86.89 |
| Mean single sequence MFE | -31.86 |
| Consensus MFE | -21.96 |
| Energy contribution | -22.60 |
| Covariance contribution | 0.64 |
| Combinations/Pair | 1.18 |
| Mean z-score | -2.32 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.806959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13153807 119 + 27905053 GCGAGUGCACACUGUACCAAAGAACUACAGCUUUGGACAGAACACAUGAGCAAUCCUUCAACAAGGAUAUUCAGUUUCAUGUAUAAAGACGUCAUUACAAUUUGUGAAAGUGAAUGCAU ((..(((....((((.((((((........))))))))))....)))..)).((((((....))))))(((((.(((((((((............)))).....))))).))))).... ( -29.70) >DroSec_CAF1 183176 119 + 1 GCGAGUGCACACUGUACCAAAGAACUACAGCUUUGGACAGAACACAUGAGCCAUCCUUCAACAAGGAUAUUCGGUUUCAUGUGGAAAGACGCCAUUACAAUUUGUGAAAGUGAAUGCAU ....(((((..((((.((((((........))))))))))..((((((((((((((((....))))))....)).))))))))......(((..((((.....))))..)))..))))) ( -36.60) >DroSim_CAF1 168456 119 + 1 GCGAGUGCACACUGUACCAAAUAUCUACAGAGUUGGACAGAACACAUGAGCCAUCCUUCAACAAGGAUAUUCAGUUUCAUGUAGAAAGACUCUAUUACAAUUUGUGCAAGUGAAUGCAU ....(((((((((((((.((((......((((((..((((((.((.((((..((((((....)))))).))))))))).))).....))))))......)))))))).))))..))))) ( -29.90) >DroEre_CAF1 174301 112 + 1 GCGAGUGCACACUGUCCUAAAGAACUACAGCUUUGGACAGAACACAUGAGCAAUCCUUCAACAAGGAUAUUCAGUUUCAUGCACCAAGCCGCCAUUACAAUUUGUGGAGGUC------- ..(.(((((..((((((.((((........))))))))))......((((..((((((....)))))).))))......))))))..(((.((((........)))).))).------- ( -32.80) >DroYak_CAF1 181904 119 + 1 GCGAGUGCACACUGUCCUAAAGAGCUACAGCUUUGCACGGAACACAUGGGCAAUCCUUCAACAAGGAUAUUCAGUUUCUUGUAUCAGGCAGCCAUGACAAUUUGUAAAAGUGAAUGCAU ....(((((((((.(((...(((((....)))))....)))...(((((.(...(((...(((((((........)))))))...)))..))))))............))))..))))) ( -30.30) >consensus GCGAGUGCACACUGUACCAAAGAACUACAGCUUUGGACAGAACACAUGAGCAAUCCUUCAACAAGGAUAUUCAGUUUCAUGUAGAAAGACGCCAUUACAAUUUGUGAAAGUGAAUGCAU ((..(((....((((.((((((........))))))))))....)))..)).((((((....))))))(((((.(((((((.....................))))))).))))).... (-21.96 = -22.60 + 0.64)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:51 2006