
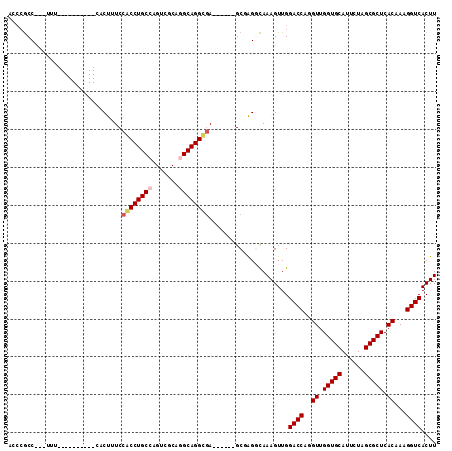
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,136,853 – 13,137,048 |
| Length | 195 |
| Max. P | 0.971719 |

| Location | 13,136,853 – 13,136,947 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -34.35 |
| Consensus MFE | -27.43 |
| Energy contribution | -27.77 |
| Covariance contribution | 0.33 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.47 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.649325 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13136853 94 + 27905053 ACCCGCC---UUU----------CACUUUCCACCUGCCAGUCGCAGGCAGGUGA------GCGAGGCAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ....(((---((.----------..((...((((((((.......)))))))))------).))))).......((((..((.(((((......))))).))...)))).... ( -35.60) >DroPse_CAF1 155545 101 + 1 UUCCGCCACCAUU------------CCAUCCGCCUGCCAGUUGCAUGCAGGCGAGGGCGAGCCGGCCAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ....(((......------------.....(((((((.........)))))))..((....)))))........((((..((.(((((......))))).))...)))).... ( -33.90) >DroEre_CAF1 156554 94 + 1 ACCCGCC---UUU----------CACUUUCCACCUGCCAGUCGCAGGCAGGUGA------GCGAGGCAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ....(((---((.----------..((...((((((((.......)))))))))------).))))).......((((..((.(((((......))))).))...)))).... ( -35.60) >DroYak_CAF1 164659 94 + 1 ACCCGCC---UUU----------CACUUUCCACCUGCCAGUCGCAGGCAGGUGA------GCGAGGCAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ....(((---((.----------..((...((((((((.......)))))))))------).))))).......((((..((.(((((......))))).))...)))).... ( -35.60) >DroAna_CAF1 155763 104 + 1 UAGAACC---UUUUCCUUCUCGGCACUUUCCACCUGCCAGCAGCAGGCAGGCAA------GCAAGGUAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ......(---(((.((((((.((......)).((((((.......))))))..)------).)))).))))...((((..((.(((((......))))).))...)))).... ( -31.50) >DroPer_CAF1 221996 101 + 1 UUCCGCCACCAUU------------CCAUCCGCCUGCCAGUUGCAUGCAGGCGAGGGCGAGCCGGCCAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ....(((......------------.....(((((((.........)))))))..((....)))))........((((..((.(((((......))))).))...)))).... ( -33.90) >consensus ACCCGCC___UUU__________CACUUUCCACCUGCCAGUCGCAGGCAGGCGA______GCGAGGCAAAGUUGGACCAGGUUGGUGCAUUCUAGCGCUCACAAAGGUCACUU ..............................((((((((.......)))))))).....................((((..((.(((((......))))).))...)))).... (-27.43 = -27.77 + 0.33)


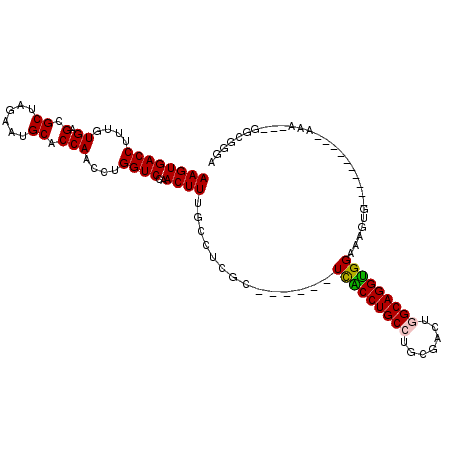
| Location | 13,136,853 – 13,136,947 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 82.13 |
| Mean single sequence MFE | -36.10 |
| Consensus MFE | -25.13 |
| Energy contribution | -24.85 |
| Covariance contribution | -0.28 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.39 |
| Structure conservation index | 0.70 |
| SVM decision value | -0.02 |
| SVM RNA-class probability | 0.523531 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13136853 94 - 27905053 AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUGCCUCGC------UCACCUGCCUGCGACUGGCAGGUGGAAAGUG----------AAA---GGCGGGU ..(.((((....((.(.((......)).)))....)))))...((((((((((------(((((((((.......))))))))...))))----------)..---)))))). ( -35.40) >DroPse_CAF1 155545 101 - 1 AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUGGCCGGCUCGCCCUCGCCUGCAUGCAACUGGCAGGCGGAUGG------------AAUGGUGGCGGAA ..((.(((...((((((((((((.((.(((.....))).))..))))))..))))))((((((((((.........))))))))..))------------...))).)).... ( -38.70) >DroEre_CAF1 156554 94 - 1 AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUGCCUCGC------UCACCUGCCUGCGACUGGCAGGUGGAAAGUG----------AAA---GGCGGGU ..(.((((....((.(.((......)).)))....)))))...((((((((((------(((((((((.......))))))))...))))----------)..---)))))). ( -35.40) >DroYak_CAF1 164659 94 - 1 AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUGCCUCGC------UCACCUGCCUGCGACUGGCAGGUGGAAAGUG----------AAA---GGCGGGU ..(.((((....((.(.((......)).)))....)))))...((((((((((------(((((((((.......))))))))...))))----------)..---)))))). ( -35.40) >DroAna_CAF1 155763 104 - 1 AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUACCUUGC------UUGCCUGCCUGCUGCUGGCAGGUGGAAAGUGCCGAGAAGGAAAA---GGUUCUA ..(.((((....((.(.((......)).)))....)))))..((((.((((.(------(((((((((.......)))))))((......))))))))).)))---)...... ( -33.00) >DroPer_CAF1 221996 101 - 1 AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUGGCCGGCUCGCCCUCGCCUGCAUGCAACUGGCAGGCGGAUGG------------AAUGGUGGCGGAA ..((.(((...((((((((((((.((.(((.....))).))..))))))..))))))((((((((((.........))))))))..))------------...))).)).... ( -38.70) >consensus AAGUGACCUUUGUGAGCGCUAGAAUGCACCAACCUGGUCCAACUUUGCCUCGC______UCACCUGCCUGCGACUGGCAGGUGGAAAGUG__________AAA___GGCGGGA ((((((((....((.(.((......)).)))....))))..))))..............(((((((((.......)))))))))............................. (-25.13 = -24.85 + -0.28)


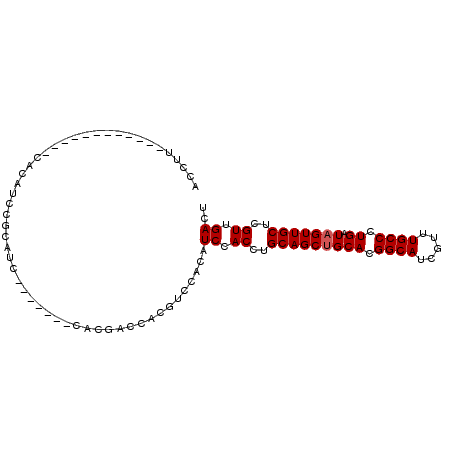
| Location | 13,136,947 – 13,137,048 |
|---|---|
| Length | 101 |
| Sequences | 4 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.62 |
| Mean single sequence MFE | -19.82 |
| Consensus MFE | -17.88 |
| Energy contribution | -18.12 |
| Covariance contribution | 0.25 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971719 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13136947 101 - 27905053 ACCUUCCCAUCCACCUUCACAUCCACAUCCACAUU-CACGACCACGUCCACAUCCACCUGCAGCUGCACGGCAUCGUUUGCCCUGAUAGUUGCUCGUUGACU ...................................-................((.((..(((((((((.((((.....)))).)).)))))))..)).)).. ( -18.90) >DroSec_CAF1 166301 83 - 1 ACCUU------------CACCUCCGCAUC-------CACGACCACGUCCACAUCCACCUGCAGCUGCACGGCAUCGUUUGCCCUGAUAGUUGCUCGUUGACU .....------------..........((-------.((((....(....)........(((((((((.((((.....)))).)).))))))))))).)).. ( -19.30) >DroSim_CAF1 151321 83 - 1 ACCUU------------CACAUCCGCAUC-------CACGACCACGUCCACAUCCACCUGCAGCUGCACGGCAUCGUUUGCCCUGAUAGUUGCUCGUUGACU .....------------..........((-------.((((....(....)........(((((((((.((((.....)))).)).))))))))))).)).. ( -19.30) >DroEre_CAF1 156648 89 - 1 ACGUGCACAGCCACCUGCACUU-------------CCACUUCCACAUCCACAUCCACCUGCAGCAGCACGGCAUCGUUUGCCCUGAUAGUUGCUCGUUGACU ..(((((........)))))..-------------.................((.((..(((((..((.((((.....)))).))...)))))..)).)).. ( -21.80) >consensus ACCUU____________CACAUCCGCAUC_______CACGACCACGUCCACAUCCACCUGCAGCUGCACGGCAUCGUUUGCCCUGAUAGUUGCUCGUUGACU ....................................................((.((..(((((((((.((((.....)))).)).)))))))..)).)).. (-17.88 = -18.12 + 0.25)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:42 2006