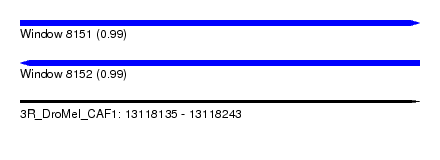
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,118,135 – 13,118,243 |
| Length | 108 |
| Max. P | 0.990257 |

| Location | 13,118,135 – 13,118,243 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -39.68 |
| Consensus MFE | -18.80 |
| Energy contribution | -21.78 |
| Covariance contribution | 2.98 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.84 |
| Structure conservation index | 0.47 |
| SVM decision value | 2.17 |
| SVM RNA-class probability | 0.989534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13118135 108 + 27905053 AAAUGUCUGCCGGAGCAUCUGGAAGAC----U-CCCAGUCUCCGUUCCCUUACUCCGACUCACUCGAAGGCUGAGCUUCUGACUCCCUGGAUACUUUCCACGGAGUCAAGAGC .....((.((((((((....(((.(((----.-....))))))))))).......(((.....)))..))).))(((..(((((((.((((.....)))).)))))))..))) ( -40.90) >DroSec_CAF1 148512 108 + 1 AAAUGUUUGCCGGAGCAUCUGGAAUAC----U-CUCAAUCUCCGGUCCCUCACCAGGACUCACUCGAAGGAUGAGCUCCUGACUCCCUGGAUACUUUCCACGGAGUCAAGAGC ........(((((((.((..(((....----)-))..)))))))))..(((...((((((((.((....)))))).))))((((((.((((.....)))).))))))..))). ( -43.20) >DroSim_CAF1 133260 108 + 1 AAAUGUCUGCCGGAGCAUCUGGAAGAC----U-CCCAAUCUCCGGUCCCUUACCAGGACUCACUCGAAGGCUGAGCUCUUGACUCCCUGGAUACUUUCCACGGAGUCAAGAGC .....((.(((((((.((..(((....----)-))..))))))(((((.......)))))........))).))((((((((((((.((((.....)))).)))))))))))) ( -47.80) >DroEre_CAF1 138611 106 + 1 AAAUGUCUGCCGCAGCAUCUGGAAGUC----U-CCCAAUCCCCGGUUCUUA-CUCCGACUCACUCGACGGCUGAGCCGCUGACUCCCUGGUUGC-UUCCACGGACUCAAGAGC ....(((((..(.((((((.((.((((----.-.........((((((...-(..(((.....)))..)...))))))..)))).)).)).)))-).)..)))))........ ( -26.40) >DroYak_CAF1 145777 108 + 1 AAAUGUCUGCCGGAGCAUCUGGAAGUC----U-CCCAAUCUCCGGUCCUUAGCUCCGGCUCACUCGAAGGCUGAGCUCCUGACUCCCUGGACGCCUUCCACGGAGUCAAGAAC ....((((((((((((..(((((..(.----.-....)..)))))......))))))))((....)))))).....((.(((((((.((((.....)))).))))))).)).. ( -41.10) >DroPer_CAF1 204791 102 + 1 AAGUGUCUGG-----AGUCUGGAGGCCGGAGUCCGGAGUCCGCGGUC---UGCCAUGUCCCAGUGCCAGUUCCAGCCACUG---CCGCGGAUACCUCAAGCGGAUUCAAGAGC .....(((((-----(.(((((...))))).))).((((((((((..---..)).(((((..(((.((((.......))))---.))))))))......)))))))).))).. ( -38.70) >consensus AAAUGUCUGCCGGAGCAUCUGGAAGAC____U_CCCAAUCUCCGGUCCCUUACCACGACUCACUCGAAGGCUGAGCUCCUGACUCCCUGGAUACUUUCCACGGAGUCAAGAGC .....((.(((((((....(((............)))..))))((((.........))))........))).))((((.(((((((.((((.....)))).))))))).)))) (-18.80 = -21.78 + 2.98)

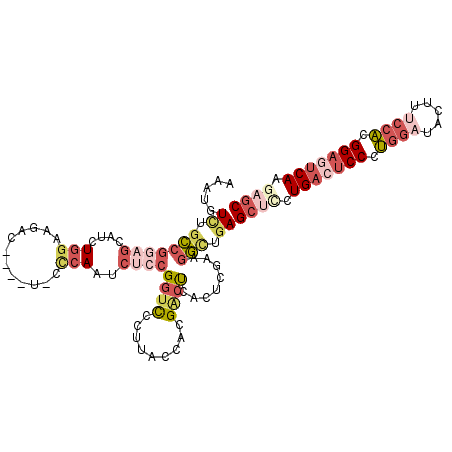
| Location | 13,118,135 – 13,118,243 |
|---|---|
| Length | 108 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 76.11 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -25.42 |
| Energy contribution | -27.23 |
| Covariance contribution | 1.81 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.66 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.20 |
| SVM RNA-class probability | 0.990257 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
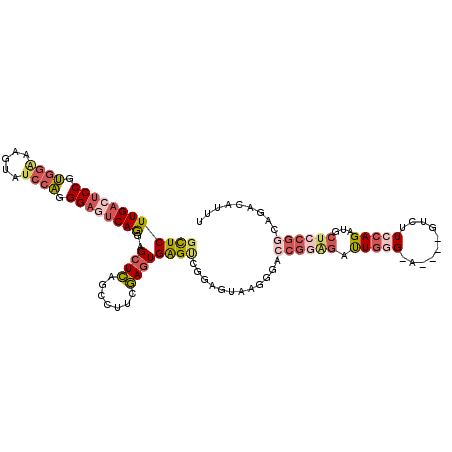
>3R_DroMel_CAF1 13118135 108 - 27905053 GCUCUUGACUCCGUGGAAAGUAUCCAGGGAGUCAGAAGCUCAGCCUUCGAGUGAGUCGGAGUAAGGGAACGGAGACUGGG-A----GUCUUCCAGAUGCUCCGGCAGACAUUU ((((.(((((((.((((.....)))).)))))))((((......))))))))..(((((((((.(....)((((((....-.----))))))....)))))))))........ ( -47.70) >DroSec_CAF1 148512 108 - 1 GCUCUUGACUCCGUGGAAAGUAUCCAGGGAGUCAGGAGCUCAUCCUUCGAGUGAGUCCUGGUGAGGGACCGGAGAUUGAG-A----GUAUUCCAGAUGCUCCGGCAAACAUUU ((((((((((((.((((.....)))).))))))))))))...(((((((...(....)...)))))))(((((((((..(-.----.....)..))).))))))......... ( -48.50) >DroSim_CAF1 133260 108 - 1 GCUCUUGACUCCGUGGAAAGUAUCCAGGGAGUCAAGAGCUCAGCCUUCGAGUGAGUCCUGGUAAGGGACCGGAGAUUGGG-A----GUCUUCCAGAUGCUCCGGCAGACAUUU ((((((((((((.((((.....)))).))))))))))))...(((.......(.(((((.....))))))((((.(((((-(----...))))))...)))))))........ ( -52.10) >DroEre_CAF1 138611 106 - 1 GCUCUUGAGUCCGUGGAA-GCAACCAGGGAGUCAGCGGCUCAGCCGUCGAGUGAGUCGGAG-UAAGAACCGGGGAUUGGG-A----GACUUCCAGAUGCUGCGGCAGACAUUU ((((.(((.(((.(((..-....))).))).)))(((((...))))).))))..((((.((-((........(((..((.-.----..)))))...)))).))))........ ( -37.10) >DroYak_CAF1 145777 108 - 1 GUUCUUGACUCCGUGGAAGGCGUCCAGGGAGUCAGGAGCUCAGCCUUCGAGUGAGCCGGAGCUAAGGACCGGAGAUUGGG-A----GACUUCCAGAUGCUCCGGCAGACAUUU ((((((((((((.((((.....)))).))))))))))))...........((..((((((((...(((.........((.-.----..)))))....))))))))..)).... ( -50.20) >DroPer_CAF1 204791 102 - 1 GCUCUUGAAUCCGCUUGAGGUAUCCGCGG---CAGUGGCUGGAACUGGCACUGGGACAUGGCA---GACCGCGGACUCCGGACUCCGGCCUCCAGACU-----CCAGACACUU ..(((.((.....((.(((((.(((((((---(((((.(.......).)))))...(......---).)))))))...(((...)))))))).))..)-----).)))..... ( -31.20) >consensus GCUCUUGACUCCGUGGAAAGUAUCCAGGGAGUCAGGAGCUCAGCCUUCGAGUGAGUCGGAGUAAGGGACCGGAGAUUGGG_A____GUCUUCCAGAUGCUCCGGCAGACAUUU ((((((((((((.((((.....)))).))))))))..((((.......))))))))............((((((.(((((..........)))))...))))))......... (-25.42 = -27.23 + 1.81)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:37 2006