| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,116,803 – 13,116,905 |
| Length | 102 |
| Max. P | 0.718625 |

| Location | 13,116,803 – 13,116,905 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 83.06 |
| Mean single sequence MFE | -39.97 |
| Consensus MFE | -25.52 |
| Energy contribution | -26.97 |
| Covariance contribution | 1.45 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.61 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.718625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
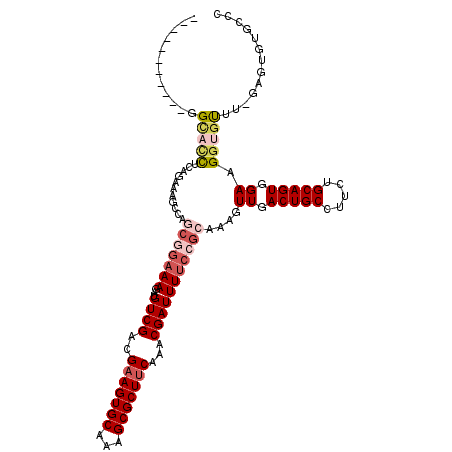
>3R_DroMel_CAF1 13116803 102 + 27905053 ------------GAAGCUCGGGAUGCAG-----AAGAUGUCGACGAAGUGCAAAGCGCUUCAAACGAUUUUCCGCAAAGUUGACUGCCUUCUGCAGUGGAAGGUGUUUCGAGUGUGCAC ------------(..(((((..((((((-----(((..(((((((((((((...)))))))...((......))....))))))...))))))))..........)..)))))...).. ( -36.60) >DroVir_CAF1 133827 107 + 1 -----------GGCACCGCAGAAAGCCAGCGGAAAGAUGUCGACGAAGUGCAAAGCGCUUCAAACGAUUUUCCGCAAAGUUGACUGCCUUUUGCAGUGGAAGGUGUUU-GAGUGUGCCU -----------((((((.((((..(((.(((((((...((((..(((((((...)))))))...)))))))))))....((.(((((.....))))).)).))).)))-).).))))). ( -44.80) >DroGri_CAF1 133715 107 + 1 -----------GGCACCAUAUAAAGCCAGCGGAAAGAUGUCGACGAAGUGCAAAGCGCUGCAAACGAUUUUCCGCAAAGUUGACUGCCUCUUGCAGUGGAAGGUGCUU-GAGUGUGCCU -----------(((((.((.(((.(((.(((((((...((((....(((((...))))).....)))))))))))....((.(((((.....))))).)).)))..))-).))))))). ( -38.70) >DroWil_CAF1 134947 107 + 1 ------------GUUUUUCGGGAAGUCAGCGGAAAGAUGUCGACGAAGUGCAAAGCGCUUCAAACGAUUUUCCGCAAAGUUGACUGCCUUCUGCAGUGGAAGGUGUUUUGAGUGUGCUC ------------((..(((((((((((((((((((...((((..(((((((...)))))))...))))))))))).....)))))(((((((.....))))))).)))))))...)).. ( -37.60) >DroMoj_CAF1 148428 107 + 1 -----------GGCACCACAGAAAGCCAGCGGAAAGAUGUCGACGAAGUGCAAAGCGCUUCAAACGAUUUUCCGCAAAGUUGACUGCUCCGUGCAGUGGAAGGUGUUU-CAGAGUGCCC -----------(((((....(((((((.(((((((...((((..(((((((...)))))))...)))))))))))....((.(((((.....))))).)).))).)))-)...))))). ( -43.50) >DroPer_CAF1 203810 118 + 1 UGCUCUGUUGUGGC-UCUCGAGGAGCAGCCCGAAAGAUGUCGACGAAGUGCAAAGCGCUUCAAACGAUUUUCCGCAAAGUUGACUGCCUUCUGCAGUGGAAGGUGUUUUGAGUGUGCAC .((((..(((.(((-((((...))).)))))))((((((((((((((((((...)))))))...((......))....)))))).(((((((.....))))))))))))))))...... ( -38.60) >consensus ___________GGCACCUCAGAAAGCCAGCGGAAAGAUGUCGACGAAGUGCAAAGCGCUUCAAACGAUUUUCCGCAAAGUUGACUGCCUUCUGCAGUGGAAGGUGUUU_GAGUGUGCCC ............(((((...........(((((((...((((..(((((((...)))))))...)))))))))))....((.(((((.....))))).)).)))))............. (-25.52 = -26.97 + 1.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:34 2006