
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,114,806 – 13,114,898 |
| Length | 92 |
| Max. P | 0.994112 |

| Location | 13,114,806 – 13,114,898 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 67.28 |
| Mean single sequence MFE | -32.15 |
| Consensus MFE | -11.31 |
| Energy contribution | -13.53 |
| Covariance contribution | 2.22 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.35 |
| SVM decision value | 2.45 |
| SVM RNA-class probability | 0.994112 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13114806 92 + 27905053 AUGGACACUCAC-UCACAUCAG-AGCCUCGU---C-GCUUGUCCCGGACUCCGUAGUCGGUAUUUUGUAUUCCGUACUACGUAGUCCGU---GUUCCGUGU ((((((((....-........(-(((.....---.-)))).....(((((.(((((((((.((.....)).))).)))))).)))))))---).))))).. ( -31.70) >DroVir_CAF1 131732 82 + 1 AUAGAUGCAGAC-ACAUUCUGA---CCG--UAUUCUGGCCAUGCCUGCCGUCGUACACGGG---C------UGAGAC---G-UGGCCGUAACGGAUCGUGU ...(((.((((.-....)))).---(((--(....((((((((((.(((..((....))))---)------.).).)---)-))))))..))))))).... ( -26.60) >DroSec_CAF1 145212 92 + 1 AUGGACACUCAC-UCACAUCGA-AGCCCCCU---C-GCUUGUUCCGGACUCCGUAGUCGGUAUUUUGUAUUCCGUACUACGUAGUCCGU---GUUCCGUGU ((((((((....-........(-(((.....---.-)))).....(((((.(((((((((.((.....)).))).)))))).)))))))---).))))).. ( -31.40) >DroSim_CAF1 129995 92 + 1 AUGGACACUCAC-UCACAUCGA-AGCCCCCU---C-GCUUGUUCCGGACUCCGUAGUCGGUAUUUUGUAUUCCGUACUACGUAGUCCGU---GUUCCGUGU ((((((((....-........(-(((.....---.-)))).....(((((.(((((((((.((.....)).))).)))))).)))))))---).))))).. ( -31.40) >DroEre_CAF1 135434 92 + 1 ACGGACACUCAC-UCACAUCGA-UGCCACGU---C-GCUUGUUCCGGACUCCGUAGUCGGUAUUUUGUAUUCCGUACUACGUAGUCCGU---GUUCCGUGU ((((((((....-..(((.(((-((...)))---)-)..)))...(((((.(((((((((.((.....)).))).)))))).)))))))---).))))).. ( -34.70) >DroAna_CAF1 135286 87 + 1 AGGGACAGACCCAUCACAGGGACUGGCCUGU---U-GGUCGCACCGGAUUCCGUGCUCCGAACUCUGCGAGCCGGAC-------UCCAU---GUUCCGUGU .((((((((((....(((((......)))))---.-)))).....(((.((((.((((((.....)).)))))))).-------))).)---))))).... ( -37.10) >consensus AUGGACACUCAC_UCACAUCGA_AGCCCCGU___C_GCUUGUUCCGGACUCCGUAGUCGGUAUUUUGUAUUCCGUACUACGUAGUCCGU___GUUCCGUGU ............................................((((((.(((((((((.((.....)).))).)))))).))))))............. (-11.31 = -13.53 + 2.22)


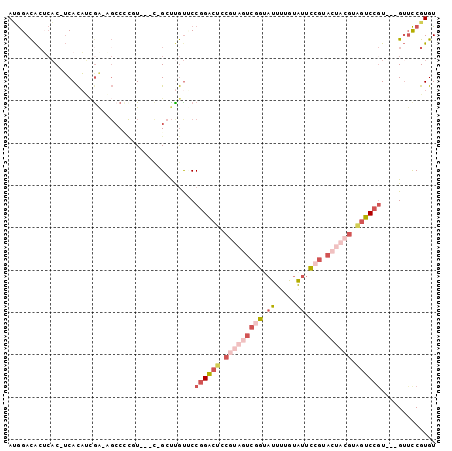
| Location | 13,114,806 – 13,114,898 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 67.28 |
| Mean single sequence MFE | -31.82 |
| Consensus MFE | -12.09 |
| Energy contribution | -14.40 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.37 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980749 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13114806 92 - 27905053 ACACGGAAC---ACGGACUACGUAGUACGGAAUACAAAAUACCGACUACGGAGUCCGGGACAAGC-G---ACGAGGCU-CUGAUGUGA-GUGAGUGUCCAU .........---.((((((.((((((.(((.((.....)).))))))))).))))))(((((..(-.---.....(((-(......))-)))..))))).. ( -33.40) >DroVir_CAF1 131732 82 - 1 ACACGAUCCGUUACGGCCA-C---GUCUCA------G---CCCGUGUACGACGGCAGGCAUGGCCAGAAUA--CGG---UCAGAAUGU-GUCUGCAUCUAU ....((.((((...(((((-.---((((..------(---((((....))..))))))).))))).....)--)))---))(((.(((-....)))))).. ( -25.80) >DroSec_CAF1 145212 92 - 1 ACACGGAAC---ACGGACUACGUAGUACGGAAUACAAAAUACCGACUACGGAGUCCGGAACAAGC-G---AGGGGGCU-UCGAUGUGA-GUGAGUGUCCAU ....(((.(---(((((((.((((((.(((.((.....)).))))))))).)))))....((..(-(---(((...))-)))...)).-....)))))).. ( -32.70) >DroSim_CAF1 129995 92 - 1 ACACGGAAC---ACGGACUACGUAGUACGGAAUACAAAAUACCGACUACGGAGUCCGGAACAAGC-G---AGGGGGCU-UCGAUGUGA-GUGAGUGUCCAU ....(((.(---(((((((.((((((.(((.((.....)).))))))))).)))))....((..(-(---(((...))-)))...)).-....)))))).. ( -32.70) >DroEre_CAF1 135434 92 - 1 ACACGGAAC---ACGGACUACGUAGUACGGAAUACAAAAUACCGACUACGGAGUCCGGAACAAGC-G---ACGUGGCA-UCGAUGUGA-GUGAGUGUCCGU ..(((((.(---(((((((.((((((.(((.((.....)).))))))))).)))))....((.((-(---.((.....-.)).)))..-.)).)))))))) ( -33.50) >DroAna_CAF1 135286 87 - 1 ACACGGAAC---AUGGA-------GUCCGGCUCGCAGAGUUCGGAGCACGGAAUCCGGUGCGACC-A---ACAGGCCAGUCCCUGUGAUGGGUCUGUCCCU ....((.((---(((((-------.((((((((..........)))).)))).))))....((((-.---(((((......)))))....))))))).)). ( -32.80) >consensus ACACGGAAC___ACGGACUACGUAGUACGGAAUACAAAAUACCGACUACGGAGUCCGGAACAAGC_G___ACGGGGCU_UCGAUGUGA_GUGAGUGUCCAU .............((((((.((((((.(((.((.....)).))))))))).))))))............................................ (-12.09 = -14.40 + 2.31)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:30 2006