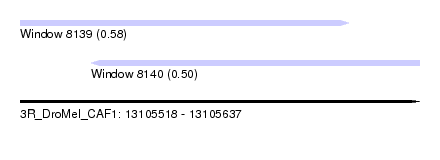
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,105,518 – 13,105,637 |
| Length | 119 |
| Max. P | 0.582674 |

| Location | 13,105,518 – 13,105,616 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 77.24 |
| Mean single sequence MFE | -29.40 |
| Consensus MFE | -15.84 |
| Energy contribution | -16.32 |
| Covariance contribution | 0.48 |
| Combinations/Pair | 1.28 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.54 |
| SVM decision value | 0.10 |
| SVM RNA-class probability | 0.582674 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
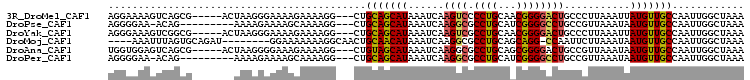
>3R_DroMel_CAF1 13105518 98 + 27905053 A---------------GCAGCAAUCUGGCGGAAAUAUUUAGCCAAUUGGCAACAUAAUUUAAGGGCAGUCCCCGUUGCAGGGGACUUGAUUUAUGCUGCAG---CCUUUUCUUUUC .---------------((((((...((((...........))))..((....)).....((((...(((((((......)))))))...))))))))))..---............ ( -31.30) >DroPse_CAF1 127830 103 + 1 A--GCAGA--------GCAGCAAUCCGGCGGAAAUAUUUAGCCAAUUGGCAACAUUAUUUAACGGCAGGCCCCGAUGCAGGCGCCUUGAUUUAUGCUGCAG---CCUUUUGCUUUU (--(((((--------((((((..(((....(((((....(((....))).....)))))..))).((((.((......)).)))).......))))))..---...))))))... ( -30.90) >DroWil_CAF1 125316 89 + 1 A---------------GCAGCAAUCUGGCGGAAAUAUUUAGCCAAUUGGCAACAUUAUUUAACGGCUCUGUCCU-------AGCCUUGAUUUAUGUUGCAG---CAUUUCUUUU-- .---------------((..((((.((((...........))))))))(((((((...((((.((((.......-------))))))))...))))))).)---).........-- ( -25.60) >DroMoj_CAF1 138022 110 + 1 GGGG----CGGUGUGCGUUGCAGACUGACGAAAGUAUUUAGCCAAUUGGCAACAUUAUUUAAGAAUUGG-CCUGCUGCAGGCGCCUUGAUUUAUGUUGCAGUUGCCUUUUUUUCC- ((((----((((.(((...))).))).((....)).....))).....(((((((...(((((.....(-((((...)))))..)))))...))))))).....)).........- ( -32.80) >DroAna_CAF1 126260 98 + 1 A---------------GCGGCAAUCUGGCGGAAAUAUUUAGCCAAUUGGCAACAUUAUUUAACGGCAGUCCCCGCUGCAGGCGCCUUGAUUUAUGCUACAG---CCUUUUCUUUCC (---------------(((..((((.((((..........(((....)))..............(((((....)))))...))))..))))..))))....---............ ( -24.90) >DroPer_CAF1 194306 103 + 1 A--GCAGA--------GCAGCAAUCCGGCGGAAAUAUUUAGCCAAUUGGCAACAUUAUUUAACGGCAGGCCCCGAUGCAGGCGCCUUGAUUUAUGCUGCAG---CCUUUUGCUUUU (--(((((--------((((((..(((....(((((....(((....))).....)))))..))).((((.((......)).)))).......))))))..---...))))))... ( -30.90) >consensus A_______________GCAGCAAUCUGGCGGAAAUAUUUAGCCAAUUGGCAACAUUAUUUAACGGCAGGCCCCGAUGCAGGCGCCUUGAUUUAUGCUGCAG___CCUUUUCUUUU_ ................(((((((((.((((..........(((....))).............((......))........))))..)))...))))))................. (-15.84 = -16.32 + 0.48)

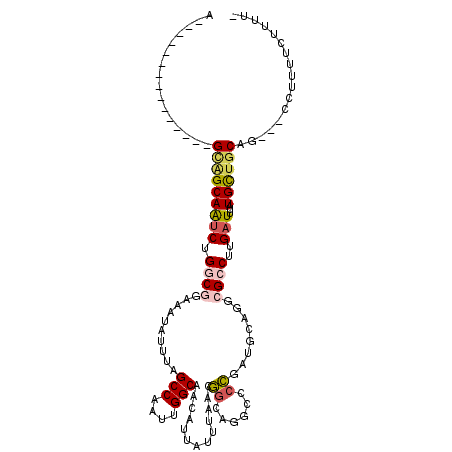
| Location | 13,105,539 – 13,105,637 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 78.55 |
| Mean single sequence MFE | -24.60 |
| Consensus MFE | -12.66 |
| Energy contribution | -12.41 |
| Covariance contribution | -0.25 |
| Combinations/Pair | 1.27 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.51 |
| SVM decision value | -0.06 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13105539 98 - 27905053 AGGAAAAGUCAGCG-----ACUAAGGGAAAAGAAAAGG---CUGCAGCAUAAAUCAAGUCCCCUGCAACGGGGACUGCCCUUAAAUUAUGUUGCCAAUUGGCUAAA ......((((((((-----((((((((....((....(---(....)).....)).(((((((......))))))).))))))......)))))....)))))... ( -30.00) >DroPse_CAF1 127856 93 - 1 AGGGGAA-ACAG---------AAAAGAAAAGCAAAAGG---CUGCAGCAUAAAUCAAGGCGCCUGCAUCGGGGCCUGCCGUUAAAUAAUGUUGCCAAUUGGCUAAA ...(...-.)..---------........(((....((---(.(((...(((....((((.((......)).))))....))).....))).))).....)))... ( -22.80) >DroYak_CAF1 130413 98 - 1 AGGGAAAGUCGGCG-----ACUAAGGGAAAAGAAAAGG---CUGCAGCAUAAAUCAAGUCGCCUGCAACGGGGACUGCCCUUAAAUUAUGUUGCCAAUUGGCUAAA ..((..(((.((((-----((((((((....((....(---(....)).....)).((((.((......)).)))).))))))......)))))).)))..))... ( -27.70) >DroMoj_CAF1 138054 93 - 1 ----AAAUUUAGUGCAGAU--------GGAAAAAAAGGCAACUGCAACAUAAAUCAAGGCGCCUGCAGCAGG-CCAAUUCUUAAAUAAUGUUGCCAAUUGGCUAAA ----...((((((.(((.(--------(.......((....))(((((((.....((((.(((((...))))-)....)))).....))))))))).))))))))) ( -21.60) >DroAna_CAF1 126281 98 - 1 UGGUGGAGUCAGCG-----ACUAAGGGGAAAGAAAAGG---CUGUAGCAUAAAUCAAGGCGCCUGCAGCGGGGACUGCCGUUAAAUAAUGUUGCCAAUUGGCUAAA ((((.((....(((-----(((((.((..........(---(((((((............).)))))))(....)..)).)))......)))))...)).)))).. ( -22.70) >DroPer_CAF1 194332 93 - 1 AGGGGAA-ACAG---------AAAAGAAAAGCAAAAGG---CUGCAGCAUAAAUCAAGGCGCCUGCAUCGGGGCCUGCCGUUAAAUAAUGUUGCCAAUUGGCUAAA ...(...-.)..---------........(((....((---(.(((...(((....((((.((......)).))))....))).....))).))).....)))... ( -22.80) >consensus AGGGAAAGUCAGCG_______UAAGGGAAAAGAAAAGG___CUGCAGCAUAAAUCAAGGCGCCUGCAACGGGGACUGCCCUUAAAUAAUGUUGCCAAUUGGCUAAA ...........................................(((((((......((((.((((...))))))))...........)))))))............ (-12.66 = -12.41 + -0.25)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:26 2006