| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,872,933 – 1,873,043 |
| Length | 110 |
| Max. P | 0.597810 |

| Location | 1,872,933 – 1,873,043 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 118 |
| Reading direction | forward |
| Mean pairwise identity | 87.70 |
| Mean single sequence MFE | -23.87 |
| Consensus MFE | -20.17 |
| Energy contribution | -19.75 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.10 |
| Mean z-score | -1.66 |
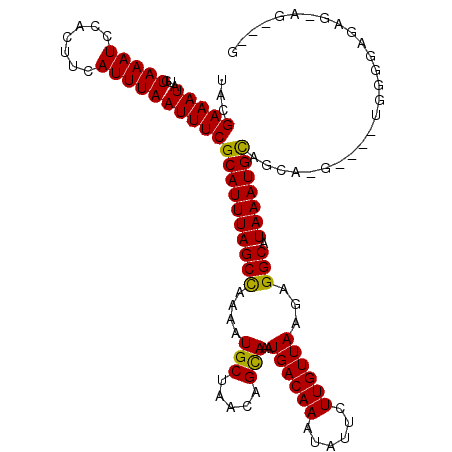
| Structure conservation index | 0.84 |
| SVM decision value | 0.13 |
| SVM RNA-class probability | 0.597810 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
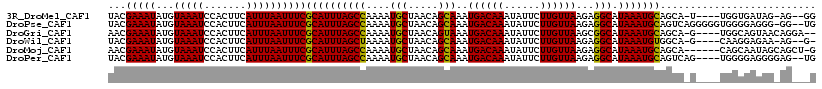
>3R_DroMel_CAF1 1872933 110 + 27905053 UACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCCAAAAUGCUAACAGCAAAUGACAAAUAUUCUUGUUAAGAGGCAUAAAUGCAGCA-U----UGGUGAUAG-AG--GG ................((.(((((((.......((((((((((....(((.....)))..((((((......))))))...))).)))))))....-.----.))))).))-.)--). ( -24.02) >DroPse_CAF1 22713 115 + 1 UACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCCAAAAUGCUAACAGCAAAUGACAAAUAUUCUUGUUAAGAGGCAUAAAUGCAGUCAGGGGGUGGGGAGGG-GG--UG ..............((((.((((((((.....(((((((((((....(((.....)))..((((((......))))))...))).))))))).).....))))))))...)-))--). ( -28.30) >DroGri_CAF1 29616 111 + 1 AACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCCAAAAUGCUAACAGUAAAUGACAAAUAUUCUUGUUAAGCGGCAUAAAUGCAGCA-G----UGGCAGUAACAGGA-- ...............(((...............((((((((((....(((.....)))..((((((......))))))...))).))))))).((.-.----..)).......)))-- ( -21.40) >DroWil_CAF1 27201 109 + 1 UACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCUAAAAUGCUAACAGCAAAUGACAAAUAUUCUUGUUAAGAGGCAUAAAUGUGGCA-G----CAAGGAGAA-AG--G- ...................((((........((((((((((((....(((.....)))..((((((......))))))...))).)))))))))..-.----...))))..-..--.- ( -18.74) >DroMoj_CAF1 29081 111 + 1 AACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCCAAAAUGCUAACAGCAAAUGACAAAUAUUCUUGUUAAGAGGCAUAAAUGCAGCA------CAGCAAUAGCAGCU-G ...(((((...(((((.......))))))))))((((((((((....(((.....)))..((((((......))))))...))).)))))))....------((((.......)))-) ( -24.60) >DroPer_CAF1 23031 112 + 1 UACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCCAAAAUGCUAACAGCAAAUGACAAAUAUUCUUGUUAAGAGGCAUAAAUGCAGUCAG----UGGGGAGGGGAG--UG ...............(((.((((..........((((((((((....(((.....)))..((((((......))))))...))).)))))))......----....))))))).--.. ( -26.15) >consensus UACGAAAUAUGUAAAUCCACUUCAUUUAAUUUCGCAUUUAGCCAAAAUGCUAACAGCAAAUGACAAAUAUUCUUGUUAAGAGGCAUAAAUGCAGCA_G____UGGGGAGAG_AG___G ...(((((...(((((.......))))))))))((((((((((....(((.....)))..((((((......))))))...))).))))))).......................... (-20.17 = -19.75 + -0.42)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:37 2006