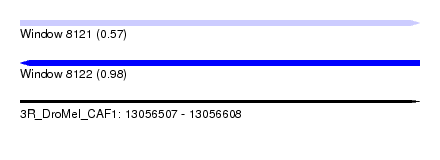
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,056,507 – 13,056,608 |
| Length | 101 |
| Max. P | 0.980812 |

| Location | 13,056,507 – 13,056,608 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -20.85 |
| Consensus MFE | -12.79 |
| Energy contribution | -13.23 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.565191 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13056507 101 + 27905053 GCUUUAAUGCUGCGGCAUAAAUCAUUUUUCCGCAAUUCGUUUCAAUUGCAUAAAAGUAUUUUAUGCACUGCCUUCUUUUGGCUGACUUU----------CAGUGUGUGUAU ((....((((((..(((((((..((((((..((((((......))))))..))))))..)))))))...(((.......))).......----------))))))..)).. ( -23.90) >DroSec_CAF1 76518 101 + 1 GCUUUAAUGCUGUAACAUAAAUCAUUUUUUCGCAAUUCGUUUCAAUUGCAUAAAAGUAUUUCAUGCACUGCCUUCUUUUGGCUGACUUU----------CAGUGUGUGUAU ((......)).............((((((..((((((......))))))..))))))....(((((((((((.......))).(.....----------)))))))))... ( -18.90) >DroEre_CAF1 76985 86 + 1 GCUGUAAUGCUGUGGCAUAAAUCAUC-UUCCGCAAUUCGUUUCAAUUGCACAAAAGUAUUUCAUGCACUGCCUUCUGUUGGCUGCCU------------------------ ((((.((((((((((......)))).-....((((((......)))))).....)))))).)).))...(((.......))).....------------------------ ( -16.50) >DroYak_CAF1 78952 101 + 1 GCUUUAAUGCUGUGGCAUAAAUCAUUUUUCCGCAAUUCGUUUCAAUUGCAUAAAAGUAUUUCAUGCACUGCCUUCUUUUGGCUGACUUU----------CAGAGUGUGUAU ......(((((.((((((((((.((((((..((((((......))))))..)))))))))).))))...(((.......))).......----------)).))))).... ( -21.40) >DroAna_CAF1 78630 105 + 1 GCUUUAAUGCUGUGGCAUAAAUCAUUUUUCCGCAAUUCGUUUCAAUUGCAAAAAAAUACUUUAUGCACUAGCUCCU-UCGGCUGCCGCUGUGGACCAC-CAGA----ACAU ........((((..(((((((..((((((..((((((......))))))..))))))..)))))))..))))(((.-.((((....)))).)))....-....----.... ( -26.40) >DroPer_CAF1 146075 107 + 1 GCUUUAAUGCUGUGGCAUAAAUCAUUUUUUCGCAAUUCGUUUCAAUUGCAUAAAAAUAUUUCAUGCUGGUGCUGCUUCUGCCUCUGCUUCUGCUUCUUCUGUUGUGU---- ........((..((((((((((.((((((..((((((......))))))..)))))))))).))))))..)).((....((....))....))..............---- ( -18.00) >consensus GCUUUAAUGCUGUGGCAUAAAUCAUUUUUCCGCAAUUCGUUUCAAUUGCAUAAAAGUAUUUCAUGCACUGCCUUCUUUUGGCUGACUUU__________CAGUGUGUGUAU ((......))....((((((((.((((((..((((((......))))))..)))))))))).)))).(.(((.......))).)........................... (-12.79 = -13.23 + 0.45)



| Location | 13,056,507 – 13,056,608 |
|---|---|
| Length | 101 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 76.90 |
| Mean single sequence MFE | -23.60 |
| Consensus MFE | -16.52 |
| Energy contribution | -16.42 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.70 |
| SVM decision value | 1.87 |
| SVM RNA-class probability | 0.980812 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13056507 101 - 27905053 AUACACACACUG----------AAAGUCAGCCAAAAGAAGGCAGUGCAUAAAAUACUUUUAUGCAAUUGAAACGAAUUGCGGAAAAAUGAUUUAUGCCGCAGCAUUAAAGC .........(((----------.......(((.......))).(.(((((((.((.((((.(((((((......))))))).)))).)).)))))))).)))......... ( -20.80) >DroSec_CAF1 76518 101 - 1 AUACACACACUG----------AAAGUCAGCCAAAAGAAGGCAGUGCAUGAAAUACUUUUAUGCAAUUGAAACGAAUUGCGAAAAAAUGAUUUAUGUUACAGCAUUAAAGC .........(((----------.......(((.......)))...(((((((.((.((((.(((((((......))))))).)))).)).)))))))..)))......... ( -18.70) >DroEre_CAF1 76985 86 - 1 ------------------------AGGCAGCCAACAGAAGGCAGUGCAUGAAAUACUUUUGUGCAAUUGAAACGAAUUGCGGAA-GAUGAUUUAUGCCACAGCAUUACAGC ------------------------..((.(((.......))).(((((((((.((.((((.(((((((......))))))).))-)))).)))))).))).))........ ( -23.70) >DroYak_CAF1 78952 101 - 1 AUACACACUCUG----------AAAGUCAGCCAAAAGAAGGCAGUGCAUGAAAUACUUUUAUGCAAUUGAAACGAAUUGCGGAAAAAUGAUUUAUGCCACAGCAUUAAAGC .........(((----------.......(((.......))).(.(((((((.((.((((.(((((((......))))))).)))).)).)))))))).)))......... ( -20.80) >DroAna_CAF1 78630 105 - 1 AUGU----UCUG-GUGGUCCACAGCGGCAGCCGA-AGGAGCUAGUGCAUAAAGUAUUUUUUUGCAAUUGAAACGAAUUGCGGAAAAAUGAUUUAUGCCACAGCAUUAAAGC ..((----(((.-.((((((.....))..)))).-.)))))..(((((((((.(((((((((((((((......))))))))))))))).)))))).)))........... ( -34.40) >DroPer_CAF1 146075 107 - 1 ----ACACAACAGAAGAAGCAGAAGCAGAGGCAGAAGCAGCACCAGCAUGAAAUAUUUUUAUGCAAUUGAAACGAAUUGCGAAAAAAUGAUUUAUGCCACAGCAUUAAAGC ----..............((....((....((....)).))....(((((((.(((((((.(((((((......))))))).))))))).)))))))....))........ ( -23.20) >consensus AUACACACACUG__________AAAGUCAGCCAAAAGAAGGCAGUGCAUGAAAUACUUUUAUGCAAUUGAAACGAAUUGCGGAAAAAUGAUUUAUGCCACAGCAUUAAAGC .............................(((.......))).(((((((((.((.((((.(((((((......))))))).)))).)).)))))).)))........... (-16.52 = -16.42 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:09 2006