
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,055,268 – 13,055,409 |
| Length | 141 |
| Max. P | 0.881269 |

| Location | 13,055,268 – 13,055,372 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -27.08 |
| Consensus MFE | -17.09 |
| Energy contribution | -19.40 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.95 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777003 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13055268 104 + 27905053 AAAUCCUCGUUUAUAAG------------CAGCUAUUAUAUUGGCCGCAACUGCUUUUUCAUAUAUGUAUUGUACAUAUAU----GUAUCUAUGGGUUCGAUUUGAGGUCGAAAGUUGGA .....(((((..(((.(------------(.((((......)))).))...........((((((((((...)))))))))----))))..)))))((((((.....))))))....... ( -24.20) >DroSec_CAF1 75302 109 + 1 AAAUCCUUGUUUAUAAGCUGCACAUUUAGCAGCUAUUAUAUUGACCGCAACUGCUUUUUCAUAUAUGUAC---AU--------ACAUAUGUAUGGGUUCGAUUUGAGGUCGAAAGUUGGA ...(((.........((((((.......))))))......((((((.(((...(...(((((((((((..---..--------..)))))))))))...)..))).)))))).....))) ( -29.00) >DroSim_CAF1 67712 117 + 1 AAAUCCUUGUUUAUAAGCUGCACAUUUAGCAGCUAUUAUAUUGGCCGCAACUGCUUUUUCAUAUAUGUAC---ACACAUAUGUACAUAUGUAUGGGUUCGAUUUGAGGUCGAAAGUUGGA ...(((.........((((((.......))))))......((((((.(((..((((....((((((((((---(......)))))))))))..)))).....))).)))))).....))) ( -32.50) >DroYak_CAF1 77760 101 + 1 AAAUCCAUGUUUGUAAGCUGCAUAUUUAGCAGCUAUUAAAUUGCCCGCAACUGCUCUUUCAUAUAUGUA-------------------UGUACUGGUUCGAUUUGAGGUCGAAAGUUGGA ...((((..((((..((((((.......)))))).((((((((.((((....)).....((((....))-------------------))....))..))))))))...))))...)))) ( -22.60) >consensus AAAUCCUUGUUUAUAAGCUGCACAUUUAGCAGCUAUUAUAUUGGCCGCAACUGCUUUUUCAUAUAUGUAC___AC__________AUAUGUAUGGGUUCGAUUUGAGGUCGAAAGUUGGA ...(((.........((((((.......))))))......((((((.(((...(...(((((((((((.................)))))))))))...)..))).)))))).....))) (-17.09 = -19.40 + 2.31)


| Location | 13,055,268 – 13,055,372 |
|---|---|
| Length | 104 |
| Sequences | 4 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.06 |
| Mean single sequence MFE | -24.40 |
| Consensus MFE | -12.64 |
| Energy contribution | -13.70 |
| Covariance contribution | 1.06 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881269 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13055268 104 - 27905053 UCCAACUUUCGACCUCAAAUCGAACCCAUAGAUAC----AUAUAUGUACAAUACAUAUAUGAAAAAGCAGUUGCGGCCAAUAUAAUAGCUG------------CUUAUAAACGAGGAUUU ............((((..................(----(((((((((...))))))))))...(((((((((............))))))------------)))......)))).... ( -24.40) >DroSec_CAF1 75302 109 - 1 UCCAACUUUCGACCUCAAAUCGAACCCAUACAUAUGU--------AU---GUACAUAUAUGAAAAAGCAGUUGCGGUCAAUAUAAUAGCUGCUAAAUGUGCAGCUUAUAAACAAGGAUUU (((.......((((................(((((((--------..---....))))))).....((....))))))..((((..((((((.......)))))))))).....)))... ( -25.60) >DroSim_CAF1 67712 117 - 1 UCCAACUUUCGACCUCAAAUCGAACCCAUACAUAUGUACAUAUGUGU---GUACAUAUAUGAAAAAGCAGUUGCGGCCAAUAUAAUAGCUGCUAAAUGUGCAGCUUAUAAACAAGGAUUU (((....(((((.......))))).((....((((((((((....))---))))))))........((....))))....((((..((((((.......)))))))))).....)))... ( -28.00) >DroYak_CAF1 77760 101 - 1 UCCAACUUUCGACCUCAAAUCGAACCAGUACA-------------------UACAUAUAUGAAAGAGCAGUUGCGGGCAAUUUAAUAGCUGCUAAAUAUGCAGCUUACAAACAUGGAUUU ((((...(((((.......)))))((....((-------------------((....)))).....((....))))..........((((((.......))))))........))))... ( -19.60) >consensus UCCAACUUUCGACCUCAAAUCGAACCCAUACAUAU__________GU___GUACAUAUAUGAAAAAGCAGUUGCGGCCAAUAUAAUAGCUGCUAAAUGUGCAGCUUAUAAACAAGGAUUU (((....(((((.......)))))...............................(((((......((....)).....)))))..((((((.......)))))).........)))... (-12.64 = -13.70 + 1.06)


| Location | 13,055,296 – 13,055,409 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -29.84 |
| Consensus MFE | -18.27 |
| Energy contribution | -20.59 |
| Covariance contribution | 2.32 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.12 |
| SVM RNA-class probability | 0.591251 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13055296 113 + 27905053 UUGGCCGCAACUGCUUUUUCAUAUAUGUAUUGUACAUAUAU----GUAUCUAUGGGUUCGAUUUGAGGUCGAAAGUUGGAGCGUGCCGAUGUGAAUAAAAUAUUCAUGGCAUCAAUC--- ..((((((...........((((((((((...)))))))))----)..((((....((((((.....))))))...))))))).)))((((((((((...)))))...)))))....--- ( -33.30) >DroSec_CAF1 75342 106 + 1 UUGACCGCAACUGCUUUUUCAUAUAUGUAC---AU--------ACAUAUGUAUGGGUUCGAUUUGAGGUCGAAAGUUGGAGCGUGCCGAUGUGAAUAAAAUAUUCAUGGCAUCAAUC--- ((((.(((..(.((((.(((((((((((..---..--------..))))))))))).(((((.....))))))))).)..)))((((...(((((((...)))))))))))))))..--- ( -28.00) >DroSim_CAF1 67752 114 + 1 UUGGCCGCAACUGCUUUUUCAUAUAUGUAC---ACACAUAUGUACAUAUGUAUGGGUUCGAUUUGAGGUCGAAAGUUGGAGCGUGCCGAUGUGAAUAAAAUAUUCAUGGCAUCAAUC--- ..((((((..(.((((((..((((((((((---(......)))))))))))..((.(((.....))).)))))))).)..))).)))((((((((((...)))))...)))))....--- ( -34.90) >DroEre_CAF1 75927 94 + 1 UUGCCCGCAACUGCUCUUCCACAUAUGAA-----------------------UUGGUUCGAUUUGAGGUCGAAAGCUGGAGCGUGCCGACGUGAAUAAAAUAUUCAUGGCAUCAAUC--- .((((.(((...(((((..(.((......-----------------------.)).((((((.....)))))).)..))))).)))....(((((((...)))))))))))......--- ( -23.50) >DroYak_CAF1 77800 101 + 1 UUGCCCGCAACUGCUCUUUCAUAUAUGUA-------------------UGUACUGGUUCGAUUUGAGGUCGAAAGUUGGAGCGUGCCGAUGUGAAUAAAAUAUUCAUGGCAUCAACGGGU ..(((((.....(((((..(((((....)-------------------))).....((((((.....)))))).)..)))))(((((...(((((((...))))))))))))...))))) ( -29.50) >consensus UUGCCCGCAACUGCUUUUUCAUAUAUGUA____A____________UAUGUAUGGGUUCGAUUUGAGGUCGAAAGUUGGAGCGUGCCGAUGUGAAUAAAAUAUUCAUGGCAUCAAUC___ ............((((((((((((((((.................)))))))))))((((((.....))))))....)))))(((((...(((((((...))))))))))))........ (-18.27 = -20.59 + 2.32)



| Location | 13,055,296 – 13,055,409 |
|---|---|
| Length | 113 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.71 |
| Mean single sequence MFE | -25.60 |
| Consensus MFE | -14.04 |
| Energy contribution | -14.20 |
| Covariance contribution | 0.16 |
| Combinations/Pair | 1.08 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.55 |
| SVM decision value | 0.41 |
| SVM RNA-class probability | 0.726945 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13055296 113 - 27905053 ---GAUUGAUGCCAUGAAUAUUUUAUUCACAUCGGCACGCUCCAACUUUCGACCUCAAAUCGAACCCAUAGAUAC----AUAUAUGUACAAUACAUAUAUGAAAAAGCAGUUGCGGCCAA ---....((((...((((((...))))))))))(((.(((..(....(((((.......)))))..........(----(((((((((...))))))))))........)..)))))).. ( -27.90) >DroSec_CAF1 75342 106 - 1 ---GAUUGAUGCCAUGAAUAUUUUAUUCACAUCGGCACGCUCCAACUUUCGACCUCAAAUCGAACCCAUACAUAUGU--------AU---GUACAUAUAUGAAAAAGCAGUUGCGGUCAA ---(((((.((((.((((((...))))))....)))).....((((((((((.......)))))......(((((((--------..---....))))))).......)))))))))).. ( -21.10) >DroSim_CAF1 67752 114 - 1 ---GAUUGAUGCCAUGAAUAUUUUAUUCACAUCGGCACGCUCCAACUUUCGACCUCAAAUCGAACCCAUACAUAUGUACAUAUGUGU---GUACAUAUAUGAAAAAGCAGUUGCGGCCAA ---....((((...((((((...))))))))))(((.(((..(....(((((.......)))))..((((..(((((((((....))---)))))))))))........)..)))))).. ( -29.60) >DroEre_CAF1 75927 94 - 1 ---GAUUGAUGCCAUGAAUAUUUUAUUCACGUCGGCACGCUCCAGCUUUCGACCUCAAAUCGAACCAA-----------------------UUCAUAUGUGGAAGAGCAGUUGCGGGCAA ---......((((.((((((...))))))....)))).((((((((((((..((.((.((.(((....-----------------------))))).)).))..))).))))).)))).. ( -24.40) >DroYak_CAF1 77800 101 - 1 ACCCGUUGAUGCCAUGAAUAUUUUAUUCACAUCGGCACGCUCCAACUUUCGACCUCAAAUCGAACCAGUACA-------------------UACAUAUAUGAAAGAGCAGUUGCGGGCAA .((((((((((...((((((...)))))))))))))..((((..((((((((.......)))))..))).((-------------------((....))))...))))......)))... ( -25.00) >consensus ___GAUUGAUGCCAUGAAUAUUUUAUUCACAUCGGCACGCUCCAACUUUCGACCUCAAAUCGAACCCAUACAUA____________U____UACAUAUAUGAAAAAGCAGUUGCGGCCAA .......((((...((((((...))))))))))(((.(((..(....(((((.......)))))...................................((......)))..)))))).. (-14.04 = -14.20 + 0.16)

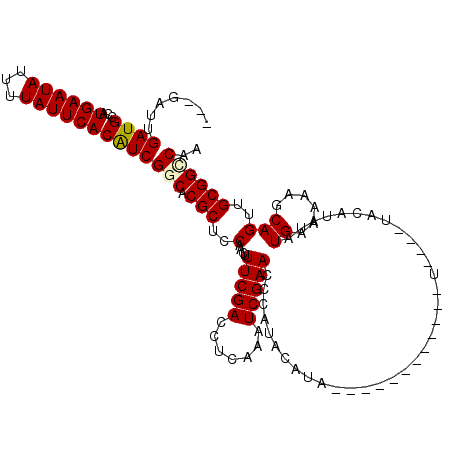

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:07 2006