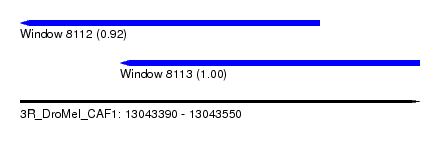
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,043,390 – 13,043,550 |
| Length | 160 |
| Max. P | 0.999943 |

| Location | 13,043,390 – 13,043,510 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.58 |
| Mean single sequence MFE | -36.72 |
| Consensus MFE | -32.60 |
| Energy contribution | -32.60 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.49 |
| Structure conservation index | 0.89 |
| SVM decision value | 1.11 |
| SVM RNA-class probability | 0.916688 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13043390 120 - 27905053 CACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUAACGGCAGGCACUUCCUUUGGAGUAUUCACAUUAGGAAUGUUGUUUCCAUGGGCAAACACUAUAGGCACAGCGGUC .((((.(((((......((((((((((...((((.....)))).....))))))))))))))).))))...........((.((((((.((.((((.......)))).)).)))))).)) ( -36.80) >DroSec_CAF1 63496 120 - 1 CACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGGAAUAUUCACAUUAGGAAUGUUGUUCCCAUGGGCAAACACAAUAGGCACAGCGGUG ((((.(....((...((((((((((((...((((.....)))).....))))))))).(((.((((((((((.......)))))))....))).)))......)))...))....))))) ( -32.80) >DroSim_CAF1 55731 119 - 1 CACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGG-AUAUUCACAUUAGGAAUGCUGUUCCCAUGGGCAAACACAAUAGGCACAGCGGUG (((((.(((((......((((((((((...((((.....)))).....))))))))))))))).)-)...............((((((.((..((........))...)).))))))))) ( -34.10) >DroEre_CAF1 63958 120 - 1 CACUUAGGAGGCAUUCUGAGUGUCUGCAAUUCGAUAUCCUCGAUCACGGCAGGCACUUCUUUUGGAAUAUUCACAUUAGGCAUGCUGUUCCCAUGGGCAAAUACCAUAGGCACAGCGGUG ((((...(((..(((((((((((((((...((((.....)))).....)))))))))).....))))).)))...........(((((.((.((((.......)))).)).))))))))) ( -40.40) >DroYak_CAF1 65314 120 - 1 CACAUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGGAAUAUUCACGUUAGGCAUGCUGUUCCCAUGGGCAAAUAGCAUAGGCACAGCGGUG (((.(.(((((......((((((((((...((((.....)))).....))))))))))))))).)........((((..((((((((((((....))..))))))))..))..))))))) ( -39.50) >consensus CACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGGAAUAUUCACAUUAGGAAUGCUGUUCCCAUGGGCAAACACAAUAGGCACAGCGGUG (((...(((((......((((((((((...((((.....)))).....)))))))))))))))...................((((((.((.((((.......)))).)).))))))))) (-32.60 = -32.60 + 0.00)



| Location | 13,043,430 – 13,043,550 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -35.78 |
| Consensus MFE | -33.38 |
| Energy contribution | -33.66 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.06 |
| Mean z-score | -2.69 |
| Structure conservation index | 0.93 |
| SVM decision value | 4.72 |
| SVM RNA-class probability | 0.999943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13043430 120 - 27905053 CCAGGGAAUUUUAGCCUUUCUUAUAGUAUUCUUACUACAGCACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUAACGGCAGGCACUUCCUUUGGAGUAUUCACAUUAGG ((((((.......(((((((...(((((....)))))........))))))).....((((((((((...((((.....)))).....)))))))))).))))))............... ( -37.60) >DroSec_CAF1 63536 120 - 1 CCAGGGAAUUUUCGCCUUUCUUCUAGUAUUUUUACUACAGCACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGGAAUAUUCACAUUAGG ((((((.......(((((((...(((((....)))))........))))))).....((((((((((...((((.....)))).....)))))))))).))))))............... ( -37.70) >DroSim_CAF1 55771 119 - 1 CCAGGGAAUUUUUGCCUUUCUUCUAGUAUUUCUACUACAGCACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGG-AUAUUCACAUUAGG ((((((......((((((((...(((((....)))))........))))))))....((((((((((...((((.....)))).....)))))))))).))))))-.............. ( -37.30) >DroEre_CAF1 63998 120 - 1 CCAGGGACUUUUCGCAUUUCUGCUAGUAUUCUUACUACAGCACUUAGGAGGCAUUCUGAGUGUCUGCAAUUCGAUAUCCUCGAUCACGGCAGGCACUUCUUUUGGAAUAUUCACAUUAGG .(((((..(((((......(((.(((((....))))))))......)))))..)))))(((((((((...((((.....)))).....))))))))).((..((((....)).))..)). ( -30.90) >DroYak_CAF1 65354 120 - 1 CCAGGGAAUCGUUGCAUUUCUGCUAGUAUUCCUACUACAGCACAUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGGAAUAUUCACGUUAGG (((((((((((.(((.((((((((((((....))))..))))...)))).)))...)))((((((((...((((.....)))).....)))))))))))).))))............... ( -35.40) >consensus CCAGGGAAUUUUCGCCUUUCUUCUAGUAUUCUUACUACAGCACUUGAGAGGCAUUUUGAGUGUCUGCAAUUCGAUAUCCUCGAUUACGGCAGGCACUUCCUUUGGAAUAUUCACAUUAGG ((((((.......(((((((...(((((....)))))........))))))).....((((((((((...((((.....)))).....)))))))))).))))))............... (-33.38 = -33.66 + 0.28)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:43:00 2006