
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,022,209 – 13,022,441 |
| Length | 232 |
| Max. P | 0.961911 |

| Location | 13,022,209 – 13,022,312 |
|---|---|
| Length | 103 |
| Sequences | 5 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 86.13 |
| Mean single sequence MFE | -33.98 |
| Consensus MFE | -25.94 |
| Energy contribution | -27.94 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.03 |
| SVM RNA-class probability | 0.546389 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13022209 103 + 27905053 -CGGGUCCGGCUCAAAAACUUGAGGUUGGGUGAUAUCACGUCAGUUGCAGGCUUCCACCGGCUUCCAUCGGCUACCACCGACUACCACCGGACUCGUUCACCGA -((((((((((((((....)))))((((((((....)))(((...((.(((((......))))).))..))).....))))).....)))))))))........ ( -36.60) >DroSec_CAF1 42256 104 + 1 CCGGGACCGGCUCAAAAACUUGAGGUUGGGUGAUAUCACGUCAGUGGCAGGCUUCCACCGGCUUCCAUCGGCUACCACCGACUACCACCGGACUCGUUCACCGA .((((.(((((((((....)))))((((((((....)))(((.((((.(((((......))))))))).))).....))))).....)))).))))........ ( -37.10) >DroSim_CAF1 34735 103 + 1 -CGGGUCCGGCUCAAAAACUUGAGGUUGGGUGAUAUCACGUCAGUUGCAGGCUUCCACCGGCUUCCAUCGGCUACCACCGACUACCACCUGACUCGUUCACCGA -((((((.(((((((....)))))((((((((....)))(((...((.(((((......))))).))..))).....))))).....)).))))))........ ( -30.50) >DroEre_CAF1 42841 82 + 1 -CGGGUCCGGCUCAA-AACUUGAGGUUGGGUGAUAUCACGUCAGUUGCAGGCUUCCA----------CCGGCU----------GCCACCGGACUCGUUUACCGA -(((((((((((((.-....))))..((((((....)))..((((((..((...)).----------.)))))----------)))))))))))))........ ( -29.30) >DroYak_CAF1 43664 93 + 1 -CGGGUCCGGCUCAAAAACUUGAGGUUGGGUGAUAUCACGUCAGUUGCAGGCUUCCAUUGGCUUCCAUUGGCU----------ACCACCGGACUCGUUCACCGA -((((((((((((((....)))))..((((((....)))((((((.(.(((((......)))))).)))))).----------.))))))))))))........ ( -36.40) >consensus _CGGGUCCGGCUCAAAAACUUGAGGUUGGGUGAUAUCACGUCAGUUGCAGGCUUCCACCGGCUUCCAUCGGCUACCACCGACUACCACCGGACUCGUUCACCGA .((((((((((((((....)))))..((((((....)))(((...((.(((((......))))).))..)))............))))))))))))........ (-25.94 = -27.94 + 2.00)


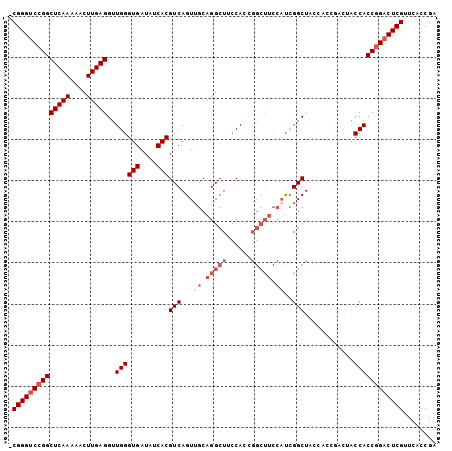
| Location | 13,022,274 – 13,022,388 |
|---|---|
| Length | 114 |
| Sequences | 4 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 88.01 |
| Mean single sequence MFE | -36.77 |
| Consensus MFE | -29.80 |
| Energy contribution | -31.30 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.43 |
| Structure conservation index | 0.81 |
| SVM decision value | 1.53 |
| SVM RNA-class probability | 0.961911 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13022274 114 + 27905053 AUCGGCUACCACCGACUACCACCGGACUCGUUCACCGACAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCUGGAGCUAUUGCUAAAGCUAAAGCCAAAGCCAA ...((((....(((........))).............(((((((((((........))))...)))))))(.(((..((((..(((....)))..))))..))))..)))).. ( -36.10) >DroSec_CAF1 42322 114 + 1 AUCGGCUACCACCGACUACCACCGGACUCGUUCACCGACAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCAGGAGCUGGAGCUACUGCUAAAGCUAAAGCCAA ...((((.((.(((........)))....((((..(..(((((((((((........))))...)))))))..)...)))).))((((..(((....)))..))))..)))).. ( -39.50) >DroSim_CAF1 34800 114 + 1 AUCGGCUACCACCGACUACCACCUGACUCGUUCACCGACAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCAGGAGCUGGAGCUACUGCUAAAGCUAAAGCCAA ...((((..............((((.(((...((.(..(((((((((((........))))...)))))))..).)))))))))((((..(((....)))..))))..)))).. ( -38.80) >DroYak_CAF1 43729 98 + 1 AUUGGCU----------ACCACCGGACUCGUUCACCGACAGUUUUUGGCACAUUUCAACCAGAUGAAACUGGAGCUGGAGCUGGAGCUGCAGCU------AUAGCUAAAGCCAA .((((((----------......(((((.(((....))))))))(((((.........((((......))))(((((.(((....))).)))))------...))))))))))) ( -32.70) >consensus AUCGGCUACCACCGACUACCACCGGACUCGUUCACCGACAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCAGGAGCUGGAGCUACUGCUAAAGCUAAAGCCAA ...((((..............((((.(((...((.(..(((((((((((........))))...)))))))..).)))))))))((((..(((....)))..))))..)))).. (-29.80 = -31.30 + 1.50)

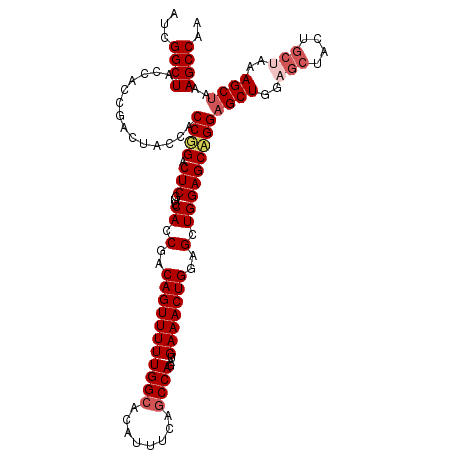

| Location | 13,022,312 – 13,022,428 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -39.84 |
| Consensus MFE | -27.58 |
| Energy contribution | -28.78 |
| Covariance contribution | 1.20 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.68 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.850919 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13022312 116 + 27905053 CAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCUGGAGCUAUUGCUAAAGCUAAAGCCAAAGCCAAAGCUGUUGCCCAAGGAGAAGCCAUUUUGGGCUUUUUGUCC (((((((.(((.........((((......)))).(((..((((..(((....)))..))))..)))....))))))))))..((((((((.(....).))))))))......... ( -41.20) >DroSec_CAF1 42360 116 + 1 CAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCAGGAGCUGGAGCUACUGCUAAAGCUAAAGCCAAAGCUGUUGCCCAAGGAGAAGCCAUUUUGGCCUUUUUGUCC (((((((.(((.........((((......))))((((..(((((.(((....))).)))))..))))...)))))))))).....(((((((..(((.....))))))))))... ( -42.70) >DroSim_CAF1 34838 116 + 1 CAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCAGGAGCUGGAGCUACUGCUAAAGCUAAAGCCAAAGCUGUUGCCCAAGGAGAAGCCAUUUUGGCCUUUUUGUCC (((((((.(((.........((((......))))((((..(((((.(((....))).)))))..))))...)))))))))).....(((((((..(((.....))))))))))... ( -42.70) >DroEre_CAF1 42923 98 + 1 CAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCUGGAGCUGGAGCU------------------AAAGCUGUUGGCCAAGGAGAAGCCAUUUUGGCCUUUCUGUCC (((((((((((........))))...)))))))(.((..(((((..(((....)))------------------..)))).)..)))..(((((.(((.....))).))))).... ( -34.10) >DroYak_CAF1 43757 110 + 1 CAGUUUUUGGCACAUUUCAACCAGAUGAAACUGGAGCUGGAGCUGGAGCUGCAGCU------AUAGCUAAAGCCAAAGCUGUUGCCCAAGGAGAAGCCAUUUUGGCCUUUUUGUCC (((((((.(((.........((((......))))(((((.(((((......)))))------.)))))...)))))))))).....(((((((..(((.....))))))))))... ( -38.50) >consensus CAGUUUUUGGCACAUUUCAGCCAGAUGAAACUGGAGCUGGAGCUGGAGCUGGAGCUA__GCUAAAGCUAAAGCCAAAGCUGUUGCCCAAGGAGAAGCCAUUUUGGCCUUUUUGUCC (((((((.(((.........((((......))))((((..(((....)))..))))...............)))))))))).....(((((((..(((.....))))))))))... (-27.58 = -28.78 + 1.20)


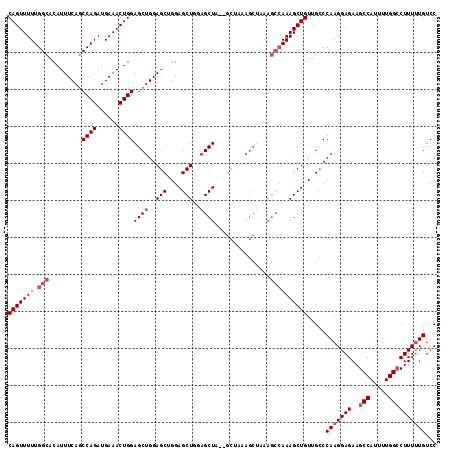
| Location | 13,022,312 – 13,022,428 |
|---|---|
| Length | 116 |
| Sequences | 5 |
| Columns | 116 |
| Reading direction | reverse |
| Mean pairwise identity | 88.65 |
| Mean single sequence MFE | -34.52 |
| Consensus MFE | -23.68 |
| Energy contribution | -26.12 |
| Covariance contribution | 2.44 |
| Combinations/Pair | 1.03 |
| Mean z-score | -2.17 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.642606 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13022312 116 - 27905053 GGACAAAAAGCCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUGGCUUUAGCUUUAGCAAUAGCUCCAGCUCCAGCUCCAGUUUCAUCUGGCUGAAAUGUGCCAAAAACUG (((....(((((......))))).)))((((((...((((...(((..(((....)))..)))...))))...)).))))...((((((....((((........)))).)))))) ( -35.30) >DroSec_CAF1 42360 116 - 1 GGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUUUAGCAGUAGCUCCAGCUCCUGCUCCAGCUCCAGUUUCAUCUGGCUGAAAUGUGCCAAAAACUG (((.....(((((.....))))).)))...((((.((..((..((...((((..(((((.(((....))).)))))..)))).(((......)))))..)).))))))........ ( -37.40) >DroSim_CAF1 34838 116 - 1 GGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUUUAGCAGUAGCUCCAGCUCCUGCUCCAGCUCCAGUUUCAUCUGGCUGAAAUGUGCCAAAAACUG (((.....(((((.....))))).)))...((((.((..((..((...((((..(((((.(((....))).)))))..)))).(((......)))))..)).))))))........ ( -37.40) >DroEre_CAF1 42923 98 - 1 GGACAGAAAGGCCAAAAUGGCUUCUCCUUGGCCAACAGCUUU------------------AGCUCCAGCUCCAGCUCCAGCUCCAGUUUCAUCUGGCUGAAAUGUGCCAAAAACUG (((((....(((((...(((((.......)))))..((((..------------------((((..(((....)))..))))..)))).....)))))....))).))........ ( -28.50) >DroYak_CAF1 43757 110 - 1 GGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUAU------AGCUGCAGCUCCAGCUCCAGCUCCAGUUUCAUCUGGUUGAAAUGUGCCAAAAACUG ((((....(((((.....))))).....(((((..(((((.((((....)))).------)))))..)).)))).))).....((((((....((((........)))).)))))) ( -34.00) >consensus GGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUUUAGC__UAGCUCCAGCUCCAGCUCCAGCUCCAGUUUCAUCUGGCUGAAAUGUGCCAAAAACUG (((.....(((((.....))))).)))..((((...((((..((((..((((........))))..))))..))))...))))((((((....((((........)))).)))))) (-23.68 = -26.12 + 2.44)


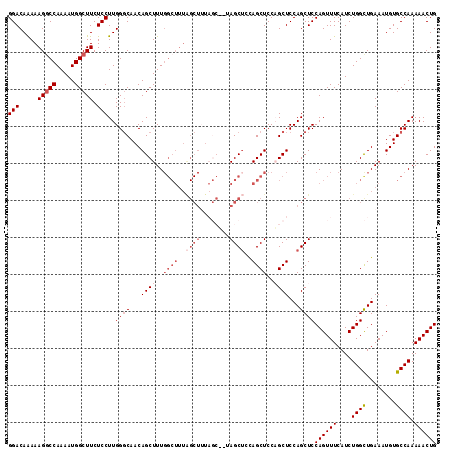
| Location | 13,022,348 – 13,022,441 |
|---|---|
| Length | 93 |
| Sequences | 5 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 80.67 |
| Mean single sequence MFE | -31.70 |
| Consensus MFE | -20.18 |
| Energy contribution | -21.88 |
| Covariance contribution | 1.70 |
| Combinations/Pair | 1.00 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.64 |
| SVM decision value | 1.43 |
| SVM RNA-class probability | 0.953493 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13022348 93 - 27905053 ----------------AAGCGGAAGUCGAGGACAAAAAGCCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUGGCUUUAGCUUUAGCAAUAGCUCCAGCUCCAG ----------------((((.((((((((((.......((((((...((.....)))))))).....)))))))))).)))).((((..(((....)))..)))).... ( -31.90) >DroSec_CAF1 42396 93 - 1 ----------------AAGCGGAAGUCGAGGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUUUAGCAGUAGCUCCAGCUCCUGCUCCAG ----------------((((.((((((((((.(((..(((((.....)))))....)))(....)..)))))))))).)))).(((((.(((....))).))))).... ( -32.30) >DroSim_CAF1 34874 93 - 1 ----------------AAGCGGAAGUCGAGGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUUUAGCAGUAGCUCCAGCUCCUGCUCCAG ----------------((((.((((((((((.(((..(((((.....)))))....)))(....)..)))))))))).)))).(((((.(((....))).))))).... ( -32.30) >DroEre_CAF1 42959 75 - 1 ----------------AAGCGGAAGUCGAGGACAGAAAGGCCAAAAUGGCUUCUCCUUGGCCAACAGCUUU------------------AGCUCCAGCUCCAGCUCCAG ----------------....(((.((((((((..((..((((.....)))))))))))))).....(((..------------------(((....)))..)))))).. ( -25.30) >DroYak_CAF1 43793 103 - 1 GAGGAAGAGGAAGGGGAAGCGGAAGUCGAGGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUAU------AGCUGCAGCUCCAGCUCCAG ..(((.....((((((((((....)).............(((.....)))))))))))((((..(((((.((((....)))).------)))))..)).))...))).. ( -36.70) >consensus ________________AAGCGGAAGUCGAGGACAAAAAGGCCAAAAUGGCUUCUCCUUGGGCAACAGCUUUGGCUUUAGCUUUAGC__UAGCUCCAGCUCCAGCUCCAG .................((((((.((((((((.....(((((.....))))).))))))).)....(((..((((...((....))...))))..)))))).))).... (-20.18 = -21.88 + 1.70)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:52 2006