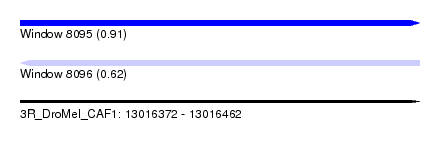
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,016,372 – 13,016,462 |
| Length | 90 |
| Max. P | 0.914648 |

| Location | 13,016,372 – 13,016,462 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -24.24 |
| Consensus MFE | -14.61 |
| Energy contribution | -17.14 |
| Covariance contribution | 2.53 |
| Combinations/Pair | 1.15 |
| Mean z-score | -2.40 |
| Structure conservation index | 0.60 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914648 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13016372 90 + 27905053 AAUUUUAUCUGGUAAC-----UAUGAAGUGGAAACCACUUCAGAUGCAAACUCUUGGCCAAACUCUUGGCUCAAGUAAACGCCAGUGAAACAAAG ..(((((.(((((...-----..((((((((...))))))))..........(((((((((....))))).)))).....))))))))))..... ( -28.60) >DroSec_CAF1 36441 85 + 1 AUUUUUUUCUGGUGAC-----UAUGAAGUGGAAACCACUUCGGAUGCAAACUUUUGGCCAAACUCUUGGCUCAAGUGAACGCCAGUGA-----AG ....(((.((((((..-----..((((((((...)))))))).......((((..((((((....)))))).))))...)))))).))-----). ( -28.00) >DroSim_CAF1 28890 90 + 1 AUUUUUUUCUGGUGAC-----UAUGAAGUGGAAACCACUUCGGAUGCAAACUUUUGGCCAAACUCUUGGCUCAAGUGAACGCCAGUGAAACAAAG ...((((.((((((..-----..((((((((...)))))))).......((((..((((((....)))))).))))...)))))).))))..... ( -28.60) >DroEre_CAF1 37014 83 + 1 AAUUUUCUCUGCUAACCAUAUUAUGACGUG------------GAUGCAAACUUUUGUCCAAACUCUUGGCUCAAGUAAGCGCCAGUGAAACAAAG ...((((.((((...((((........)))------------)..((..((((..(.((((....)))))..))))..))).))).))))..... ( -16.40) >DroYak_CAF1 37682 95 + 1 GGUUUUUUCUAGUAACCAUAUUAUGUAGUGGAAACCACUUCGGAUGCAAACUUUUGGCCAAACUGUUGGUUCAAGUAAACGGCAGUGAAACAAAG ((((((..(((((((.....)))).)))..))))))..((((..(((.....(((((((((....)))).)))))......))).))))...... ( -19.60) >consensus AAUUUUUUCUGGUAAC_____UAUGAAGUGGAAACCACUUCGGAUGCAAACUUUUGGCCAAACUCUUGGCUCAAGUAAACGCCAGUGAAACAAAG ...((((.(((((..........((((((((...))))))))..........(((((((((....))))).)))).....))))).))))..... (-14.61 = -17.14 + 2.53)

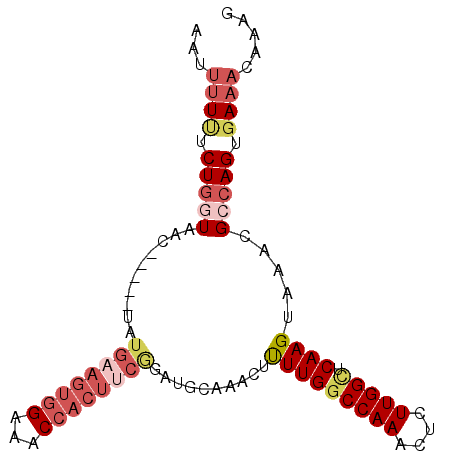
| Location | 13,016,372 – 13,016,462 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 95 |
| Reading direction | reverse |
| Mean pairwise identity | 81.60 |
| Mean single sequence MFE | -21.48 |
| Consensus MFE | -15.97 |
| Energy contribution | -17.74 |
| Covariance contribution | 1.77 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.58 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.17 |
| SVM RNA-class probability | 0.617374 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
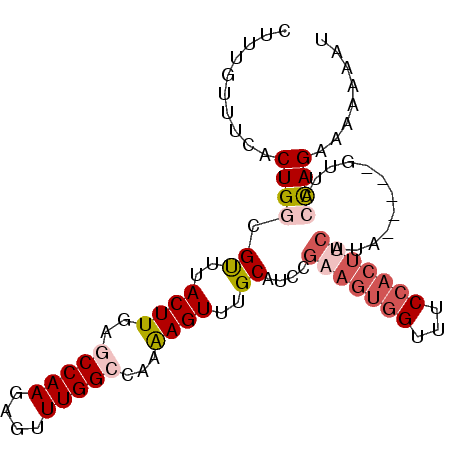
>3R_DroMel_CAF1 13016372 90 - 27905053 CUUUGUUUCACUGGCGUUUACUUGAGCCAAGAGUUUGGCCAAGAGUUUGCAUCUGAAGUGGUUUCCACUUCAUA-----GUUACCAGAUAAAAUU ..........((((.((..((((..(((((....)))))...))))..))...((((((((...))))))))..-----....))))........ ( -26.00) >DroSec_CAF1 36441 85 - 1 CU-----UCACUGGCGUUCACUUGAGCCAAGAGUUUGGCCAAAAGUUUGCAUCCGAAGUGGUUUCCACUUCAUA-----GUCACCAGAAAAAAAU ..-----...((((.((..((((..(((((....)))))...))))..))....(((((((...)))))))...-----....))))........ ( -24.50) >DroSim_CAF1 28890 90 - 1 CUUUGUUUCACUGGCGUUCACUUGAGCCAAGAGUUUGGCCAAAAGUUUGCAUCCGAAGUGGUUUCCACUUCAUA-----GUCACCAGAAAAAAAU ..........((((.((..((((..(((((....)))))...))))..))....(((((((...)))))))...-----....))))........ ( -24.50) >DroEre_CAF1 37014 83 - 1 CUUUGUUUCACUGGCGCUUACUUGAGCCAAGAGUUUGGACAAAAGUUUGCAUC------------CACGUCAUAAUAUGGUUAGCAGAGAAAAUU (((((((..(((((.((..((((...((((....))))....))))..))..)------------))(((......))))).)))))))...... ( -16.00) >DroYak_CAF1 37682 95 - 1 CUUUGUUUCACUGCCGUUUACUUGAACCAACAGUUUGGCCAAAAGUUUGCAUCCGAAGUGGUUUCCACUACAUAAUAUGGUUACUAGAAAAAACC .....((((...(((((..((((...((((....))))....))))..))....(.(((((...))))).).......))).....))))..... ( -16.40) >consensus CUUUGUUUCACUGGCGUUUACUUGAGCCAAGAGUUUGGCCAAAAGUUUGCAUCCGAAGUGGUUUCCACUUCAUA_____GUUACCAGAAAAAAAU ..........((((.((..((((..(((((....)))))...))))..))....(((((((...)))))))............))))........ (-15.97 = -17.74 + 1.77)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:44 2006