
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,015,847 – 13,015,973 |
| Length | 126 |
| Max. P | 0.805660 |

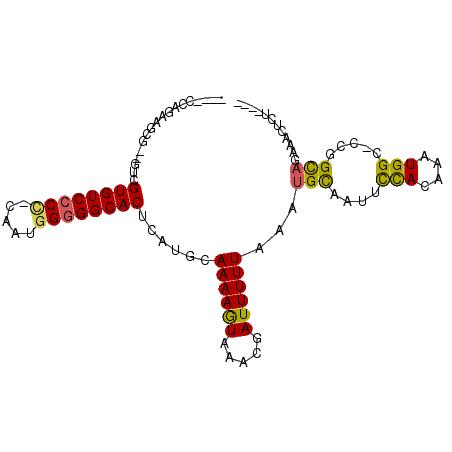
| Location | 13,015,847 – 13,015,937 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 80.96 |
| Mean single sequence MFE | -25.77 |
| Consensus MFE | -15.07 |
| Energy contribution | -14.77 |
| Covariance contribution | -0.30 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.58 |
| SVM decision value | 0.39 |
| SVM RNA-class probability | 0.716110 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13015847 90 - 27905053 ----CCAGAAGCG--GUCGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGUAAUUCCACAAAUGGC-CCGGCAGAAACUUU---- ----......(((--(((((((((((-....)))))))).(((..((((((.....))))))..))).............)))-)..)).........---- ( -23.70) >DroSec_CAF1 35914 92 - 1 ----CCAGAAGCGGGGUCGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCAAUUCCACAAAUGGC-CCGGCAGAAACUCU---- ----......((.(((((((((((((-....))))))))...((((....((((.....))))..))))...........)))-)).)).........---- ( -31.90) >DroSim_CAF1 28363 92 - 1 ----CCAAAAGCGGGGUCGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCAAUUCCACAAAUGGC-CUGGCAGAAACUCU---- ----......((.(((((((((((((-....))))))))...((((....((((.....))))..))))...........)))-)).)).........---- ( -30.30) >DroEre_CAF1 36437 82 - 1 ------------C--GAAGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCAAUUCCACAAAUGGC-CCGGCAGAAGCUCA---- ------------.--((.((((((((-....))))))))))..((((((((.....))))))...(((....(((....))).-...)))...))...---- ( -23.00) >DroYak_CAF1 37110 95 - 1 ----CCAGGAGCG--GUCGUGUCCCCUCGGUCGUGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCAAUUCCACAAAUGGC-CCGGUAGAACCUCUCUCU ----...((((.(--((....((..((.((((((((....(((..((((((.....))))))..))).....))))....)))-).))..))))).)))).. ( -20.40) >DroAna_CAF1 36731 92 - 1 CCUUCCAUGAAAA--UGCGUGUCCCU-CGAUGGGGGCACUCAUGCAAAAAUAAAAGAUUUUUAAAUGCAAUUUUAUAAAUGGCUCCGGCCAACAC------- ......((((((.--(((((((((((-....))))))))......((((((.....))))))....))).))))))...((((....))))....------- ( -25.30) >consensus ____CCAGAAGCG__GUCGUGUCCCC_CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCAAUUCCACAAAUGGC_CCGGCAGAAACUCU____ ..................((((((((.....))))))))......((((((.....))))))...(((....(((....))).....)))............ (-15.07 = -14.77 + -0.30)


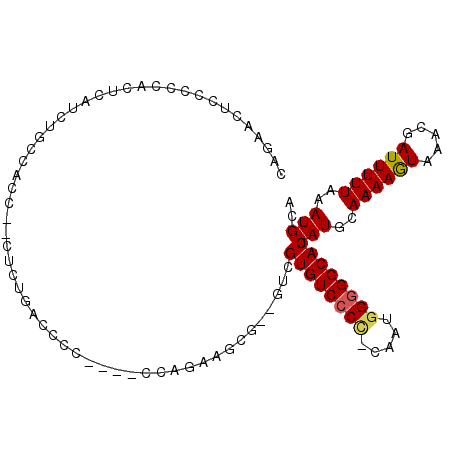
| Location | 13,015,875 – 13,015,973 |
|---|---|
| Length | 98 |
| Sequences | 6 |
| Columns | 107 |
| Reading direction | reverse |
| Mean pairwise identity | 76.41 |
| Mean single sequence MFE | -22.67 |
| Consensus MFE | -13.88 |
| Energy contribution | -13.97 |
| Covariance contribution | 0.08 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.61 |
| Structure conservation index | 0.61 |
| SVM decision value | 0.63 |
| SVM RNA-class probability | 0.805660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13015875 98 - 27905053 CAGAACUCCCCCACUCAUCAGCCACC--CUCUGACCCC----CCAGAAGCG--GUCGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGUA ....................(((.(.--.((((.....----.)))).).)--)).((((((((-....)))))))).(((..((((((.....))))))..))).. ( -23.10) >DroSec_CAF1 35942 100 - 1 CAGAACUCCCCCACUCAUCUGCCACC--CUCUGACCCC----CCAGAAGCGGGGUCGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCA .((((.((....(((....(((....--....((((((----.(....).))))))((((((((-....))))))))....)))..)))....)).))))....... ( -29.10) >DroSim_CAF1 28391 100 - 1 CAGAACUCCCCCACUCAUCUGCCACC--CUCUGACCCC----CCAAAAGCGGGGUCGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCA .((((.((....(((....(((....--....((((((----.(....).))))))((((((((-....))))))))....)))..)))....)).))))....... ( -29.10) >DroEre_CAF1 36465 77 - 1 CAGAACUCCCCCUCUCAGCUACC---------------------------C--GAAGUGUCCCC-CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCA .................((....---------------------------.--((.((((((((-....))))))))))....((((((.....))))))....)). ( -18.20) >DroYak_CAF1 37142 98 - 1 CAGAACUUCCCCUCCCAUCUACCGCC--CCCUCACCC-----CCAGGAGCG--GUCGUGUCCCCUCGGUCGUGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCA .......................(((--.(((.....-----..))).(((--..((........))..)))))).....((((....((((.....))))..)))) ( -15.50) >DroAna_CAF1 36757 100 - 1 CAGAGCACCUCU----AUCUGCCCCAAUUUUCGUCCCCCCUUCCAUGAAAA--UGCGUGUCCCU-CGAUGGGGGCACUCAUGCAAAAAUAAAAGAUUUUUAAAUGCA ....(((.....----((((((....((((((((..........)))))))--)))((((((((-....))))))))...............)))).......))). ( -21.00) >consensus CAGAACUCCCCCACUCAUCUGCCACC__CUCUGACCCC____CCAGAAGCG__GUCGUGUCCCC_CAAUGGGGGCACUCAUGCAAAAGUAAACGAUUUUUAAAUGCA ........................................................((((((((.....)))))))).(((..((((((.....))))))..))).. (-13.88 = -13.97 + 0.08)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:41 2006