| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 13,008,327 – 13,008,421 |
| Length | 94 |
| Max. P | 0.884355 |

| Location | 13,008,327 – 13,008,421 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -25.56 |
| Consensus MFE | -19.33 |
| Energy contribution | -19.50 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.20 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.75 |
| SVM RNA-class probability | 0.840643 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
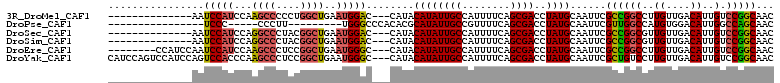
>3R_DroMel_CAF1 13008327 94 + 27905053 --------------AAUCCAUCCAAGCCCCCUGGCUGAAUGGAC---CAUACAUAUUGCCAUUUUCAGCGACCUAUGCAAUUCGCCGGCCUUGUUGACAUUGUCCGGCAAC --------------..(((((...((((....))))..))))).---....((((((((........))))..))))......((((((..((....))..).)))))... ( -25.10) >DroPse_CAF1 36319 81 + 1 ----------------UCCC-----CCCUU---------UGGGCCCACACGCAUAUUGCCGUUUUCAGCGACCUAUGCAAUUCGUUGGCCAUGUGGACAUUGGCCAGCAAC ----------------...(-----((...---------.))).......(((((((((.(....).))))..))))).....((((((((((....)).))))))))... ( -25.20) >DroSec_CAF1 28452 94 + 1 --------------AAUCCAUCCAGGCCCUACGGCUGAAUGGAC---CAUACAUAUUGCCAUUUUCAGCGACCUAUGCAAUUCGCCGGCGUUGUUGACAUUGUCCGGCAAC --------------..(((((...((((....))))..))))).---....((((((((........))))..))))......(((((((.((....)).)).)))))... ( -25.90) >DroSim_CAF1 20817 94 + 1 --------------AAUCCAUCCAGGCCCUACGGCUGAAUGGAC---CAUACAUAUUGCCAUUUUCAGCGACCUAUGCAAUUCGCCGGCGUUGUUGACAUUGUCCGGCAAC --------------..(((((...((((....))))..))))).---....((((((((........))))..))))......(((((((.((....)).)).)))))... ( -25.90) >DroEre_CAF1 29060 100 + 1 --------CCAUCCAAUCCAUCCAAGCCCUCCGGCUGAAUGGGC---CAUACAUAUUGCCAUUUUCAGCGACCUAUGCAAUUCGCCGGCCUUGUUGACAUUGUCCGGCAAC --------.................((......((((((..(((---.((....)).)))...)))))).......)).....((((((..((....))..).)))))... ( -24.72) >DroYak_CAF1 29631 108 + 1 CAUCCAGUCCAUCCAGUCCACCCAAGCCCUCCGGCUGAAUGGGC---CAUACAUAUUGCCAUUUUCAGCGACCUAUGCAAUUCGCUGUCCUUGUUGACAUUGUCCGGCAAC ......((((((.(((((..............))))).))))))---........(((((.....((((((..........))))))........(((...))).))))). ( -26.54) >consensus ______________AAUCCAUCCAAGCCCUCCGGCUGAAUGGAC___CAUACAUAUUGCCAUUUUCAGCGACCUAUGCAAUUCGCCGGCCUUGUUGACAUUGUCCGGCAAC ................(((((...((((....))))..)))))........((((((((........))))..))))......((((((..((....))..).)))))... (-19.33 = -19.50 + 0.17)


| Location | 13,008,327 – 13,008,421 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 80.45 |
| Mean single sequence MFE | -32.68 |
| Consensus MFE | -25.31 |
| Energy contribution | -26.03 |
| Covariance contribution | 0.73 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.90 |
| Structure conservation index | 0.77 |
| SVM decision value | 0.93 |
| SVM RNA-class probability | 0.884355 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 13008327 94 - 27905053 GUUGCCGGACAAUGUCAACAAGGCCGGCGAAUUGCAUAGGUCGCUGAAAAUGGCAAUAUGUAUG---GUCCAUUCAGCCAGGGGGCUUGGAUGGAUU-------------- .(((((((.(..((....))..))))))))..((((((....(((......)))..)))))).(---((((((((((((....))).))))))))))-------------- ( -35.30) >DroPse_CAF1 36319 81 - 1 GUUGCUGGCCAAUGUCCACAUGGCCAACGAAUUGCAUAGGUCGCUGAAAACGGCAAUAUGCGUGUGGGCCCA---------AAGGG-----GGGA---------------- .(((.((((((.((....)))))))).)))...(((((....((((....))))..))))).......(((.---------...))-----)...---------------- ( -25.30) >DroSec_CAF1 28452 94 - 1 GUUGCCGGACAAUGUCAACAACGCCGGCGAAUUGCAUAGGUCGCUGAAAAUGGCAAUAUGUAUG---GUCCAUUCAGCCGUAGGGCCUGGAUGGAUU-------------- .(((((((....((....))...)))))))..((((((....(((......)))..)))))).(---((((((((((((....)).)))))))))))-------------- ( -34.60) >DroSim_CAF1 20817 94 - 1 GUUGCCGGACAAUGUCAACAACGCCGGCGAAUUGCAUAGGUCGCUGAAAAUGGCAAUAUGUAUG---GUCCAUUCAGCCGUAGGGCCUGGAUGGAUU-------------- .(((((((....((....))...)))))))..((((((....(((......)))..)))))).(---((((((((((((....)).)))))))))))-------------- ( -34.60) >DroEre_CAF1 29060 100 - 1 GUUGCCGGACAAUGUCAACAAGGCCGGCGAAUUGCAUAGGUCGCUGAAAAUGGCAAUAUGUAUG---GCCCAUUCAGCCGGAGGGCUUGGAUGGAUUGGAUGG-------- ....(((..(((((((((((.((((.((.....))...))))(((......)))....))).))---))((((((((((....))).)))))))))))..)))-------- ( -34.60) >DroYak_CAF1 29631 108 - 1 GUUGCCGGACAAUGUCAACAAGGACAGCGAAUUGCAUAGGUCGCUGAAAAUGGCAAUAUGUAUG---GCCCAUUCAGCCGGAGGGCUUGGGUGGACUGGAUGGACUGGAUG ((((((((((...)))........((((((..........))))))....))))))).......---..((((((((((....))).))))))).(..(.....)..)... ( -31.70) >consensus GUUGCCGGACAAUGUCAACAAGGCCGGCGAAUUGCAUAGGUCGCUGAAAAUGGCAAUAUGUAUG___GCCCAUUCAGCCGGAGGGCUUGGAUGGAUU______________ .(((((((.(..((....))..))))))))..((((((....(((......)))..)))))).......((((((((((....))).)))))))................. (-25.31 = -26.03 + 0.73)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:37 2006