
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,986,354 – 12,986,482 |
| Length | 128 |
| Max. P | 0.643519 |

| Location | 12,986,354 – 12,986,465 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 77.91 |
| Mean single sequence MFE | -20.87 |
| Consensus MFE | -11.63 |
| Energy contribution | -11.63 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.72 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.629528 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
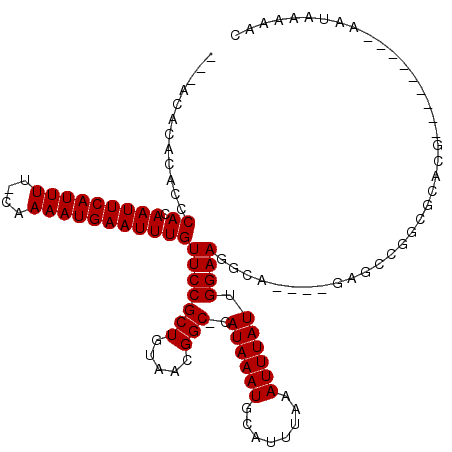
>3R_DroMel_CAF1 12986354 111 - 27905053 ---ACACAUACUCACAAUUCAUUUU-GAAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGAAGGCA----AAGUCUGCGUAUGCCACCACCAAACAACAAC ---...(((((....((((((((..-...))))))))..((((((((...))))-.((((((........)))))).)))).(((----.....)))))))).................. ( -17.70) >DroPse_CAF1 14392 102 - 1 ---ACACACACCCACAAUUCAUUUU-CAAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGAAGGCA----GCGCCGGGGCACG---------AAUAAAAAC ---.......(((((((((((((..-...)))))).))))..((((((.....(-(((((((........))))).)))...)))----)))..))).....---------......... ( -24.40) >DroWil_CAF1 7599 100 - 1 CACACACAUAUACACAAUUCAUUUUCCAAAAUGAAUUUGUUCCGCUGUAACGGCCCAUAAAUGCAUUUAAAUUUAUUGGAAUGCG----AG----------------ACCAAAAAAAAAA ...............(((((((((....))))))))).((..(((........((.((((((........)))))).))...)))----..----------------))........... ( -14.70) >DroMoj_CAF1 5761 99 - 1 ---AA--ACAUGCACAAUUCAUUUU-CGAAAUGAAUUUGUUCCGCUGUAAGGGC-CAUAAAUGCAUUUAAAUUUAUUGGAAUGCA----CGACAGCAGCAG----------UAUAAGAAG ---..--((.(((((((((((((..-...)))))).))))...(((((..(..(-(((((((........))))).)))..)...----..))))).))))----------)........ ( -20.40) >DroAna_CAF1 6777 105 - 1 ---ACACAUACUCACAAUUCAUUUU-CGAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGAAGGCAGCCUGAGUCUGCGUAAAC----------CAACAAC ---((.((.(((((.((((((((..-...))))))))......(((((.....(-(((((((........))))).)))...))))).))))).)).))....----------....... ( -23.60) >DroPer_CAF1 80323 102 - 1 ---GCACACACCCACAAUUCAUUUU-CAAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGAAGGCA----GCGCCGGGGCACG---------AAUAAAAAC ---.......(((((((((((((..-...)))))).))))..((((((.....(-(((((((........))))).)))...)))----)))..))).....---------......... ( -24.40) >consensus ___ACACACACCCACAAUUCAUUUU_CAAAAUGAAUUUGUUCCGCUGUAACGGC_CAUAAAUGCAUUUAAAUUUAUUGGAAGGCA____GAGCCGGCGCACG_________AAUAAAAAC ............((.(((((((((....)))))))))))(((((((.....)))..((((((........)))))).))))....................................... (-11.63 = -11.63 + 0.00)


| Location | 12,986,390 – 12,986,482 |
|---|---|
| Length | 92 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.24 |
| Mean single sequence MFE | -14.79 |
| Consensus MFE | -10.13 |
| Energy contribution | -10.13 |
| Covariance contribution | 0.00 |
| Combinations/Pair | 1.00 |
| Mean z-score | -1.07 |
| Structure conservation index | 0.69 |
| SVM decision value | 0.23 |
| SVM RNA-class probability | 0.643519 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12986390 92 - 27905053 --------AGCAUGCAGAAGG--------------AAAA----ACACAUACUCACAAUUCAUUUU-GAAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGA --------....(((((..((--------------((..----.........((.((((((((..-...)))))))))))))).)))))....(-(((((((........))))).))). ( -15.00) >DroVir_CAF1 6614 96 - 1 --------CAUGUGGAUAAAA--AAAUAAA-----AAUA----AA--ACAUACACAAUUCAUUUU-CCAAAUGAAUUUGUUCCGCUGUAAGGGC-CAUAAAUGCAU-UAAAUUUAUUGGA --------...(((((.....--.......-----....----..--....(((.((((((((..-...))))))))))))))))........(-(((((((....-...))))).))). ( -13.24) >DroPse_CAF1 14419 106 - 1 --------AACAUCCCGAAAAAAAACCAGGCCCCGCACC----ACACACACCCACAAUUCAUUUU-CAAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGA --------.................(((((((..(((.(----....(((.....((((((((..-...)))))))))))...).)))...)))-)((((((........))))))))). ( -16.30) >DroWil_CAF1 7619 106 - 1 CUAUGAGAAAGAUGAAAAAAA--------------AAAAACACACACAUAUACACAAUUCAUUUUCCAAAAUGAAUUUGUUCCGCUGUAACGGCCCAUAAAUGCAUUUAAAUUUAUUGGA .....................--------------................(((.(((((((((....)))))))))))).(((......)))((.((((((........)))))).)). ( -12.70) >DroMoj_CAF1 5787 88 - 1 --------UACGUGAAUAAAA--AA------------------AA--ACAUGCACAAUUCAUUUU-CGAAAUGAAUUUGUUCCGCUGUAAGGGC-CAUAAAUGCAUUUAAAUUUAUUGGA --------....(.((((((.--..------------------..--..(((((.((((((((..-...))))))))((..((.......))..-))....))))).....)))))).). ( -12.80) >DroPer_CAF1 80350 104 - 1 --------AACAUCCCGAAAA-AAACCAGGCCCC-CACC----GCACACACCCACAAUUCAUUUU-CAAAAUGAAUUUGUUCCGCUGUAACGGC-CAUAAAUGCAUUUAAAUUUAUUGGA --------.............-...(((((((..-.((.----((..(((.....((((((((..-...)))))))))))...)).))...)))-)((((((........))))))))). ( -18.70) >consensus ________AACAUGCAGAAAA__AA__________AAAA____ACACACACACACAAUUCAUUUU_CAAAAUGAAUUUGUUCCGCUGUAACGGC_CAUAAAUGCAUUUAAAUUUAUUGGA .....................................................(((((((((((....))))))).))))((((((.....)))..((((((........)))))).))) (-10.13 = -10.13 + 0.00)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:29 2006