
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,965,774 – 12,966,094 |
| Length | 320 |
| Max. P | 0.998551 |

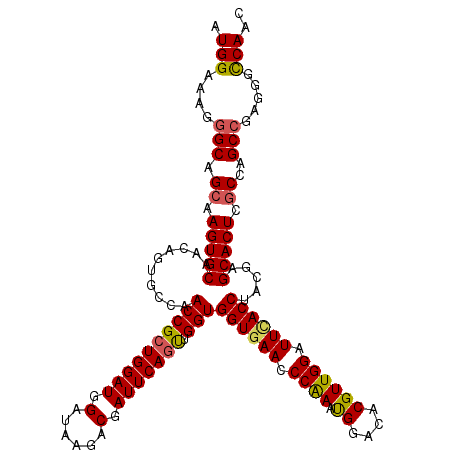
| Location | 12,965,774 – 12,965,894 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -42.02 |
| Consensus MFE | -39.02 |
| Energy contribution | -38.90 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.00 |
| Structure conservation index | 0.93 |
| SVM decision value | 3.14 |
| SVM RNA-class probability | 0.998551 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12965774 120 + 27905053 AUGGAAAGGGCAGCAAGUGCAACAGUGCCAACCGCUGGAUGGAUAAGACAAUUCAAUUGGUGGUAAACCCAAAUGGACACGUUGGAUUCACCUAUGAGCACUCACCAGCCGAGGGCCAAC .(((....(((.((....))...((((((((((((..(.(((((......))))).)..)))))....((((.((....)))))).........)).))))).....))).....))).. ( -33.30) >DroSec_CAF1 5803 120 + 1 AUGGAAAGGGCAGCAAGUGCAACAGUGCCAACCGCUGGAUGGAUAAAACGAUUCAGUUGGUGGUGAACCCAAAUGGACACGUUGGAUUCACCUACGAGCACUCGCCAGCCGAGGGCCAAC .(((....(((.((.(((((......((((...(((((((.(......).)))))))))))((((((.((((.((....)))))).)))))).....))))).))..))).....))).. ( -42.70) >DroSim_CAF1 6007 120 + 1 AUGGAAAGGGCAGCAAGUGCAACAGUGCCAACCGCUGGAUGGAUAAAACGAUUCAGUUGGUGGUGAACCCAAAUGGACACGUUGGAUUCACCUACGAGCACUCGCCAGCCGAGGGCCAAC .(((....(((.((.(((((......((((...(((((((.(......).)))))))))))((((((.((((.((....)))))).)))))).....))))).))..))).....))).. ( -42.70) >DroEre_CAF1 5781 120 + 1 AUGGAAAGGGCAGCCAGUGCAACAGUGGCAACCGCUGGAUGGAUAAGACGAUUCAGCUGGUGGUGAACCCGAAUGGAAACGUUGGAUUCACCUACGAGCACUCGCCGGCCGAGGGUCAAC ..((..((.((.((((.((...)).)))).((((((((((.(......).))))))).)))((((((.((.((((....)))))).)))))).....)).))..))((((...))))... ( -46.70) >DroYak_CAF1 5943 120 + 1 AUGGAAAGGGCAGCCAGUGCAACAGCGCCAACCGCUGGAUGGAUAAGACGAUUCAGUUGGUGGUGAACCCGAACGGAAACGUUGGAUUUACCUACGAGCACUCGCCGGCGGAGGGUCAAC ........(((..((.((((....))))...(((((((.(((((......)))))(((.((((((((.((.((((....)))))).)))))).)).))).....))))))).)))))... ( -44.70) >consensus AUGGAAAGGGCAGCAAGUGCAACAGUGCCAACCGCUGGAUGGAUAAGACGAUUCAGUUGGUGGUGAACCCAAAUGGACACGUUGGAUUCACCUACGAGCACUCGCCAGCCGAGGGCCAAC .(((....(((.((.(((((..........((((((((((.(......).))))))).)))((((((.((((.((....)))))).)))))).....))))).))..))).....))).. (-39.02 = -38.90 + -0.12)


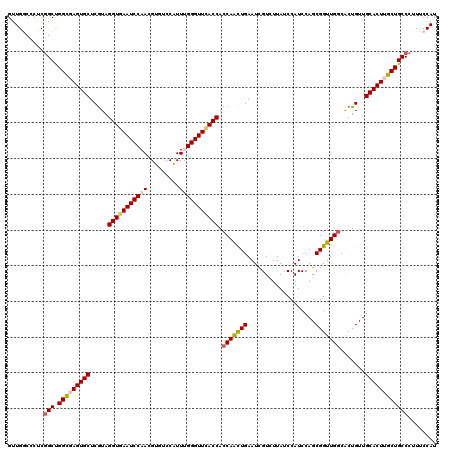
| Location | 12,965,774 – 12,965,894 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.33 |
| Mean single sequence MFE | -40.13 |
| Consensus MFE | -35.47 |
| Energy contribution | -36.27 |
| Covariance contribution | 0.80 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.71 |
| Structure conservation index | 0.88 |
| SVM decision value | 1.74 |
| SVM RNA-class probability | 0.975098 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12965774 120 - 27905053 GUUGGCCCUCGGCUGGUGAGUGCUCAUAGGUGAAUCCAACGUGUCCAUUUGGGUUUACCACCAAUUGAAUUGUCUUAUCCAUCCAGCGGUUGGCACUGUUGCACUUGCUGCCCUUUCCAU ..(((.....(((.((..(((((.....(((((((((((.........))))))))))).(((((((...((.......)).....))))))).......)))))..)))))....))). ( -38.60) >DroSec_CAF1 5803 120 - 1 GUUGGCCCUCGGCUGGCGAGUGCUCGUAGGUGAAUCCAACGUGUCCAUUUGGGUUCACCACCAACUGAAUCGUUUUAUCCAUCCAGCGGUUGGCACUGUUGCACUUGCUGCCCUUUCCAU ..(((.....(((.(((((((((.....(((((((((((.........))))))))))).(((((((...................))))))).......))))))))))))....))). ( -44.51) >DroSim_CAF1 6007 120 - 1 GUUGGCCCUCGGCUGGCGAGUGCUCGUAGGUGAAUCCAACGUGUCCAUUUGGGUUCACCACCAACUGAAUCGUUUUAUCCAUCCAGCGGUUGGCACUGUUGCACUUGCUGCCCUUUCCAU ..(((.....(((.(((((((((.....(((((((((((.........))))))))))).(((((((...................))))))).......))))))))))))....))). ( -44.51) >DroEre_CAF1 5781 120 - 1 GUUGACCCUCGGCCGGCGAGUGCUCGUAGGUGAAUCCAACGUUUCCAUUCGGGUUCACCACCAGCUGAAUCGUCUUAUCCAUCCAGCGGUUGCCACUGUUGCACUGGCUGCCCUUUCCAU .........((((((((((..(((.((.(((((((((.............))))))))))).)))....))))).........(((((((....)))))))....))))).......... ( -39.02) >DroYak_CAF1 5943 120 - 1 GUUGACCCUCCGCCGGCGAGUGCUCGUAGGUAAAUCCAACGUUUCCGUUCGGGUUCACCACCAACUGAAUCGUCUUAUCCAUCCAGCGGUUGGCGCUGUUGCACUGGCUGCCCUUUCCAU .............((((.(((((.....(((.(((((((((....)))).))))).))).(((((((...................))))))).......))))).)))).......... ( -34.01) >consensus GUUGGCCCUCGGCUGGCGAGUGCUCGUAGGUGAAUCCAACGUGUCCAUUUGGGUUCACCACCAACUGAAUCGUCUUAUCCAUCCAGCGGUUGGCACUGUUGCACUUGCUGCCCUUUCCAU ..........(((.(((((((((.....(((((((((((.........))))))))))).(((((((...................))))))).......))))))))))))........ (-35.47 = -36.27 + 0.80)


| Location | 12,965,814 – 12,965,934 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.83 |
| Mean single sequence MFE | -32.40 |
| Consensus MFE | -29.98 |
| Energy contribution | -30.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.93 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.904938 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12965814 120 - 27905053 UUCAUCUUCUUCACUACAUAGUCCAUCAUCAUUGCGAUCGGUUGGCCCUCGGCUGGUGAGUGCUCAUAGGUGAAUCCAACGUGUCCAUUUGGGUUUACCACCAAUUGAAUUGUCUUAUCC .................................((((((((((((..........((((....)))).(((((((((((.........))))))))))).))))))).)))))....... ( -26.30) >DroSec_CAF1 5843 120 - 1 UUCAUCUUCUUCACCACAUAGUCCAUCAUCAUGGCGAUCGGUUGGCCCUCGGCUGGCGAGUGCUCGUAGGUGAAUCCAACGUGUCCAUUUGGGUUCACCACCAACUGAAUCGUUUUAUCC ................................((((((((((((((.((((.....)))).)).....(((((((((((.........)))))))))))..)))))).))))))...... ( -35.60) >DroSim_CAF1 6047 120 - 1 UUCAUCUUCUUCACCACAUAGUCCAUCAUCAUGGCGAUCGGUUGGCCCUCGGCUGGCGAGUGCUCGUAGGUGAAUCCAACGUGUCCAUUUGGGUUCACCACCAACUGAAUCGUUUUAUCC ................................((((((((((((((.((((.....)))).)).....(((((((((((.........)))))))))))..)))))).))))))...... ( -35.60) >DroEre_CAF1 5821 120 - 1 UUCAUCUUCUUCACCACAUAGUCCAUCAUCAUGGCGAUCGGUUGACCCUCGGCCGGCGAGUGCUCGUAGGUGAAUCCAACGUUUCCAUUCGGGUUCACCACCAGCUGAAUCGUCUUAUCC ....................(.((((....)))))....(((((.....)))))(((((..(((.((.(((((((((.............))))))))))).)))....)))))...... ( -33.82) >DroYak_CAF1 5983 120 - 1 UUCAUCUUCUUCACCACAUAGUCCAUCAUCAUGGCGAUCGGUUGACCCUCCGCCGGCGAGUGCUCGUAGGUAAAUCCAACGUUUCCGUUCGGGUUCACCACCAACUGAAUCGUCUUAUCC ................................((((((((((((...........((((....)))).(((.(((((((((....)))).))))).)))..)))))).))))))...... ( -30.70) >consensus UUCAUCUUCUUCACCACAUAGUCCAUCAUCAUGGCGAUCGGUUGGCCCUCGGCUGGCGAGUGCUCGUAGGUGAAUCCAACGUGUCCAUUUGGGUUCACCACCAACUGAAUCGUCUUAUCC ................................(((((((((((((..........((((....)))).(((((((((((.........))))))))))).))))))).))))))...... (-29.98 = -30.10 + 0.12)

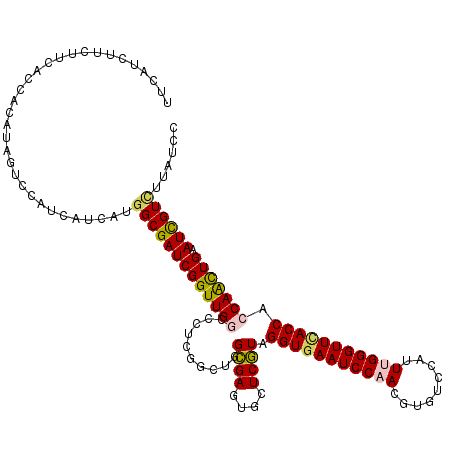

| Location | 12,965,934 – 12,966,054 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 91.50 |
| Mean single sequence MFE | -34.08 |
| Consensus MFE | -28.20 |
| Energy contribution | -28.64 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.34 |
| Structure conservation index | 0.83 |
| SVM decision value | 0.11 |
| SVM RNA-class probability | 0.590314 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12965934 120 + 27905053 GGAUGACCCUAGUUAUGGGGAAUGCGGAUCGGAAAACUUUAUUCCAGCGAAGAAAAUAAAGUUCUGUGAUGUAAGCAAAUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGA ..(((.(((((....)))))..(((.....((((.......)))).((((....(((...((((..((.((....))....))..))))....))).....)))))))......)))... ( -26.40) >DroSec_CAF1 5963 120 + 1 GGAUGACCCUAGUUACGGGGAAUGCGGAUCGGAGGACUUUAUUCCAGCGAAGAAAAUAAUGUUCUGUGAUCUAAGCAAGUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGA (.((..(((.......)))..)).)((((((.(((((.(((((...........))))).))))).)))))).(((...(((.....)))...))).....(((((........))))). ( -30.00) >DroSim_CAF1 6167 120 + 1 GGAUGACCCUAGUUACGGGGAAUGCGGAUCGGAGGACUUUAUUCCAGCGAAGAAAAUAAAGUUCUGUGAUCUAAGCAAGUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGA (.((..(((.......)))..)).)((((((.(((((((((((...........))))))))))).)))))).(((...(((.....)))...))).....(((((........))))). ( -34.10) >DroEre_CAF1 5941 120 + 1 GGAUGACCCCAGCUACGGGGAAUGCGGAUCGGAGGCCUUUAUUCCGGCACGGAAAAUAAAGUUCUGCGAUGUUAGCAAGUGCGUGGAGCAAUGGCUGAUAAUCGCGCAGAAGAACGUGGA ..(((.((((......))))..(((((((((.((..((((((((((...))))..))))))..)).)))(((((((...(((.....)))...))))))).)).))))......)))... ( -41.30) >DroYak_CAF1 6103 120 + 1 GGAUGACCCGAGUUACGGGGAAUGCGGAUCGGAGGACUUUACUCCAGCGCGGAAAAUAAAGUUCUGCGAGGUGAGCAAGUGCGUGGAGCAGUGGCUGAUAAUCGCGCAAAAGAACGUGGA (.((..((((.....))))..)).)...(((.(((((((((.(((.....)))...))))))))).))).((.(((.(.(((.....))).).))).))..(((((........))))). ( -38.60) >consensus GGAUGACCCUAGUUACGGGGAAUGCGGAUCGGAGGACUUUAUUCCAGCGAAGAAAAUAAAGUUCUGUGAUGUAAGCAAGUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGA ......((((......))))..(((..((((.((((((((((((.......))..)))))))))).))))....)))..((((((((((....))).....)))))))............ (-28.20 = -28.64 + 0.44)


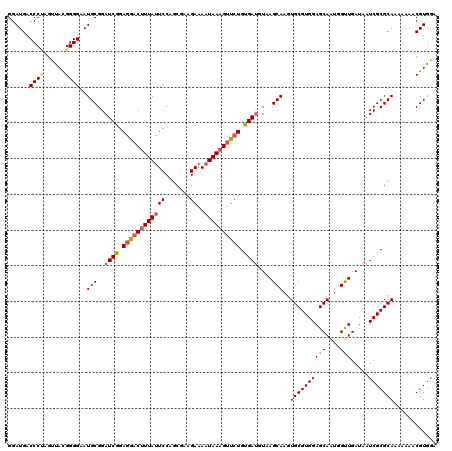
| Location | 12,965,974 – 12,966,094 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 92.25 |
| Mean single sequence MFE | -34.26 |
| Consensus MFE | -29.72 |
| Energy contribution | -28.80 |
| Covariance contribution | -0.92 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.96 |
| Structure conservation index | 0.87 |
| SVM decision value | 2.16 |
| SVM RNA-class probability | 0.989356 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12965974 120 + 27905053 AUUCCAGCGAAGAAAAUAAAGUUCUGUGAUGUAAGCAAAUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGACAAACUUGUCCAGGACCUGCAAAUGAAGGUUCUUAAGUUC ......((..((((.......))))(((((...(((...(((.....)))...)))....))))))).....(((.((((((....))))))((((((........))))))....))). ( -29.00) >DroSec_CAF1 6003 120 + 1 AUUCCAGCGAAGAAAAUAAUGUUCUGUGAUCUAAGCAAGUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGACAAACUUGUCCAGGAUCUGCAAAUGAAGGUUCUUAAGUUC ......((((....(((..(((((..((..((.....))..))..)))))...))).....)))).......(((.((((((....))))))((((((........))))))....))). ( -27.40) >DroSim_CAF1 6207 120 + 1 AUUCCAGCGAAGAAAAUAAAGUUCUGUGAUCUAAGCAAGUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGACAAACUUGUCCAGGAUCUGCAAAUGAAGGUUCUUAAGUUC ......((..((((.......))))(((((...(((...(((.....)))...)))....))))))).....(((.((((((....))))))((((((........))))))....))). ( -26.40) >DroEre_CAF1 5981 120 + 1 AUUCCGGCACGGAAAAUAAAGUUCUGCGAUGUUAGCAAGUGCGUGGAGCAAUGGCUGAUAAUCGCGCAGAAGAACGUGGACAAACUUGUCCAGGACCUGCAAAUGAAGGUUCUCAAGUUC .(((((...))))).......(((((((.(((((((...(((.....)))...)))))))....)))))))((((.((((((....))))))((((((........))))))....)))) ( -44.50) >DroYak_CAF1 6143 120 + 1 ACUCCAGCGCGGAAAAUAAAGUUCUGCGAGGUGAGCAAGUGCGUGGAGCAGUGGCUGAUAAUCGCGCAAAAGAACGUGGACAAACUUGUCCGGGACCUUCAAAUGAAGGUUCUUAAGUUC .((((..(((((((.......)))))))..).)))....((((((((((....))).....)))))))...((((..(((((....)))))(((((((((....)))))))))...)))) ( -44.00) >consensus AUUCCAGCGAAGAAAAUAAAGUUCUGUGAUGUAAGCAAGUGCGUGGAGCAAUGGUUGAUAAUCGCGCAAAAAAACGUGGACAAACUUGUCCAGGACCUGCAAAUGAAGGUUCUUAAGUUC ......((..((((.......))))(((((...(((...(((.....)))...)))....))))))).....(((.((((((....))))))((((((........))))))....))). (-29.72 = -28.80 + -0.92)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:22 2006