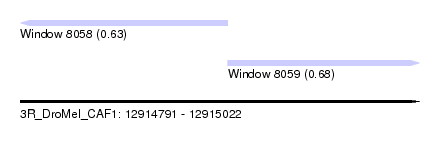
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,914,791 – 12,915,022 |
| Length | 231 |
| Max. P | 0.684826 |

| Location | 12,914,791 – 12,914,911 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 82.67 |
| Mean single sequence MFE | -44.47 |
| Consensus MFE | -30.11 |
| Energy contribution | -30.12 |
| Covariance contribution | 0.01 |
| Combinations/Pair | 1.46 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.68 |
| SVM decision value | 0.20 |
| SVM RNA-class probability | 0.630625 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
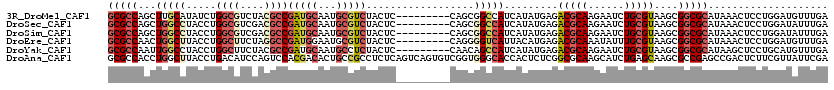
>3R_DroMel_CAF1 12914791 120 - 27905053 UCCCUCUCACGUCGGCGAAGCAACACGGCCUGCAUGUGGUGACGAUGCUCCUGCUCGUCACCAUAGGCGUACUUUGCCCGCAACUCGUCUCGUUUGUGAGAGUGUUUACGGUACAUCUUA ...((((((((..(((((.(....((((..(((.(((((((((((.((....)))))))))))))((((.....)))).)))..)))))))))))))))))).................. ( -45.30) >DroPse_CAF1 7459 120 - 1 UCCCUCUCACGACGCCGAUGUAGCAAAGCCUGCAUGUGAUGGCGGUGCUCCUGCUCAUCGUCGUAGGCGUACUUGGCACGUAGCUCGUCGCGCUUGCACGAGUGUGUGCGAUACAUUUUG ....((.(((.(((((((((.((((.(((((((........)))).)))..)))))))))..((((((((((..(((.....))).)).))))))))....))))))).))......... ( -41.60) >DroEre_CAF1 3244 120 - 1 UCCCUCUCACGUCGGCGAAGCAACACCGCCUGCAUGUGGUGACGAUGCUCCUGCUCGUCAUCGUAGGCGUACUUUGCCCGCAGCUCGUCCCGCUUGUGAGAGUGUUUACGGUACAUCUUA ...(((((((((.(((((((......(((((((....((((((((.((....)))))))))))))))))..))))))).))(((.......))).))))))).................. ( -47.30) >DroYak_CAF1 3274 120 - 1 UCCCUCUCACGUCGACGAAGCAGCACAGCCUGCAUGUGGUGACGAUGCUCCUGCUCGUCACCGUAGGCAUACUUGGCCCGCAACUCGUCCCGUUUGUGAGAGUGUUUACGGUACAUCUUA ...((((((((..(((((.((((......))))....((((((((.((....))))))))))(..(((.......)))..)...))))).....)))))))).................. ( -44.40) >DroAna_CAF1 3191 120 - 1 UCUUUCUCGCGCCGUCGGUGCAGCAGGGCUUGCAUGUGGUGGCGAUGCUCUUGUUCGUCAUCAUAAGCGUACUUGGCCCUCAGCUCGUCCCGCUUGUGGGAAUGUUUUCUGUACAUCUUU ...((((((((..(.(((..(((((((((((((.(((((((((((.((....))))))))))))).))).....))))))..))).)..)))).)))))))).................. ( -44.10) >DroPer_CAF1 3141 120 - 1 UCCCUCUCACGACGCCGAUGUAGCAAAGCCUGCAUGUGAUGGCGGUGCUCCUGUUCAUCGUCGUAGGCGUACUUGGCACGCAGCUCGUCGCGCUUGCACGAGUGUGUGCGAUACAUUUUG .......((.((((((.((((((......))))))...(((((((((........))))))))).))))).).))(((((((.(((((.((....))))))))))))))........... ( -44.10) >consensus UCCCUCUCACGUCGCCGAAGCAGCACAGCCUGCAUGUGGUGACGAUGCUCCUGCUCGUCAUCGUAGGCGUACUUGGCCCGCAGCUCGUCCCGCUUGUGAGAGUGUUUACGGUACAUCUUA ...((((((((.((((((.((.((...((((((.(((((((((((.((....))))))))))))).))).....)))..)).)))))..)))..)))))))).................. (-30.11 = -30.12 + 0.01)



| Location | 12,914,911 – 12,915,022 |
|---|---|
| Length | 111 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.59 |
| Mean single sequence MFE | -39.47 |
| Consensus MFE | -23.63 |
| Energy contribution | -24.97 |
| Covariance contribution | 1.34 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.76 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.684826 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12914911 111 + 27905053 GCGCCAGCUUGCAUAUCUGGCGUCUACGCCGAUGCAAUGCGUCUACUC---------CAGCGGCCAUCAUAUGAGACGCAAGAAUCUGCGUAAGCGGCGCAUAAACUCCUGGAUGUUUGA (((((.((((((.((((.((((....))))))))...(((((((.(..---------...............)))))))).........))))))))))).(((((........))))). ( -39.63) >DroSec_CAF1 11501 111 + 1 GCGCCAGCUGGCCUACCUGGCGUCGACGCCGAUGCAAUGCGUCUACUC---------CAGCGGCCAUCAUAUGAGACGCAAGAAUCUGCGUAAGCGGCGCAUAAACUCCUGGAUAUUUGA (((((.(((((((...((((.((.(((((.........))))).)).)---------))).))))..........(((((......))))).)))))))).................... ( -42.40) >DroSim_CAF1 10214 111 + 1 GCGCCAGCUGGCCUACCUGGCGUCGACGCCGAUGCAAUGCGUCUACUC---------CAGCGGCCAUCAUAUGAGACGCAAGAAUCUGCGUAAGCGGCGCAUAAACUCCUGGAUAUUUGA (((((.(((((((...((((.((.(((((.........))))).)).)---------))).))))..........(((((......))))).)))))))).................... ( -42.40) >DroEre_CAF1 3364 111 + 1 GCGCCAACUGGCUUACCUGGCUUCUAGGCCGAUGGAAUGCGUCUACUC---------CAGGGGUCAUUACAUGAGACGCAAAUAUUUGCGUAAGCGGCGCAUAAACUCCUGGAUGUUUGA (((((....)))...(((((((....)))))..))...)).((.((((---------(((((((.......((..((((((....))))))...))........))))))))).))..)) ( -35.56) >DroYak_CAF1 3394 111 + 1 GCGCCAAUUGGCCUACCUGGCUUCUACGCCGAUGCAAUGCCUCUACUC---------CAACAGCCAUCAUAUGAGACGCAAGAAUCUGCGUAAGCGGCGCAUAAGCUCCUGCAUGUUUGA (((((....((((.....))))..(((((.(((....(((.(((.(..---------...............)))).)))...))).)))))...))))).(((((........))))). ( -30.83) >DroAna_CAF1 3311 120 + 1 GCGCCACCUGGCUUACCUGACAUCCAGUCCACGACACUGCCGCCUCUCAGUCAGUGUCGGUGGGCACCACUCUCGGCGCAAGCAUCUGAGCAAGCGCCGAGCCGACUCUUCGUUAUUCGA ..(((....))).....((((.....(((((((((((((.(........).))))))).))))))......((((((((..((......))..))))))))..........))))..... ( -46.00) >consensus GCGCCAGCUGGCCUACCUGGCGUCUACGCCGAUGCAAUGCGUCUACUC_________CAGCGGCCAUCAUAUGAGACGCAAGAAUCUGCGUAAGCGGCGCAUAAACUCCUGGAUAUUUGA (((((...(((((.....((((....))))(((((...)))))..................))))).........(((((......)))))....))))).................... (-23.63 = -24.97 + 1.34)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:09 2006