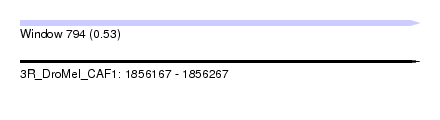
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,856,167 – 1,856,267 |
| Length | 100 |
| Max. P | 0.530039 |

| Location | 1,856,167 – 1,856,267 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 81.40 |
| Mean single sequence MFE | -22.72 |
| Consensus MFE | -13.99 |
| Energy contribution | -14.35 |
| Covariance contribution | 0.36 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.66 |
| Structure conservation index | 0.62 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.530039 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1856167 100 + 27905053 AUGUAUUUAAAAUUGACAAAUGGUUAGCAGCAUUUGUUGUUGGCAUUGCUGUUAGCUUUGUUAAGAUUUAUGUUAAGGC-AAACAAAAAUUCACAAACA------------UG ((((........(((((((..(((((((((((..(((.....))).))))))))))))))))))(((((.((((.....-.)))).))))).....)))------------). ( -25.60) >DroPse_CAF1 8524 112 + 1 AUGUAUUUAAAAUUGACAAAUGGUUACCAGCAUUAGUUGCUGGCAUUUCUGUUAGCUUUGUUAAGAUUUAUGUUCAGGC-AAACAAAUAUACACAAACACACGAGUACGAGUC .(((((((....(((((((..(((((((((((.....)))))).........))))))))))))......((((.....-.)))).................))))))).... ( -20.20) >DroEre_CAF1 13964 100 + 1 AUGUAUUUAAAAUUGACAAAUGGUUAGCAGCAUUUGUUGUUGGCAUUGCUGUUAGCUUUGUUAAGAUUUAUGUUGAGGC-AAACAGAAACGCACAAACA------------UG ............(((((((..(((((((((((..(((.....))).))))))))))))))))))....((((((...((-..........))...))))------------)) ( -26.30) >DroWil_CAF1 8184 88 + 1 AUGUAUUUAAAAUUGACAAAUGGUUAGCAGAAUUUCUUCUUGGUAUUGUUGUUAGCUUUGUUAAGAUUUAUGUUGAGGACAUAUCAAU------------------------- ((((.(((((..(((((((..(((((((((.....((....)).....))))))))))))))))........))))).))))......------------------------- ( -15.80) >DroAna_CAF1 7733 100 + 1 AUGUAUUUAAAAUUGACAAAUGGUUAGCAGCAUUUGUUGUUGGCAUGUCUGUUAGCUUUGUUAAGAUUUAUGUUGAGGC-AAACAAAAACAUACACGCA------------UA .(((((........((((....(((((((((....))))))))).))))((((.((((((.............))))))-.)))).....)))))....------------.. ( -21.72) >DroPer_CAF1 8565 112 + 1 AUGUAUUUAAAAUUGACAAAUGGUUAGCAGCAUUUGUUGCUGGCAUUGCUGUUAGCUUUGUUAAGAUUUAUGUUCAGGC-AAACAAAUAUACACAAACACACGAGUACGAGUC .(((((((....(((((((..(((((((((((..(((.....))).))))))))))))))))))......((((.....-.)))).................))))))).... ( -26.70) >consensus AUGUAUUUAAAAUUGACAAAUGGUUAGCAGCAUUUGUUGUUGGCAUUGCUGUUAGCUUUGUUAAGAUUUAUGUUGAGGC_AAACAAAAAUACACAAACA____________UC .......((((.(((((((..(((((((((....(((.....)))...))))))))))))))))..))))........................................... (-13.99 = -14.35 + 0.36)

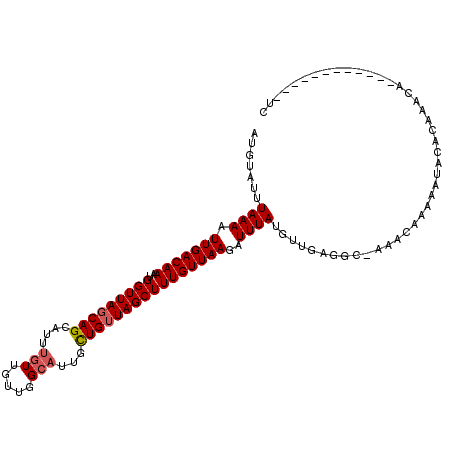
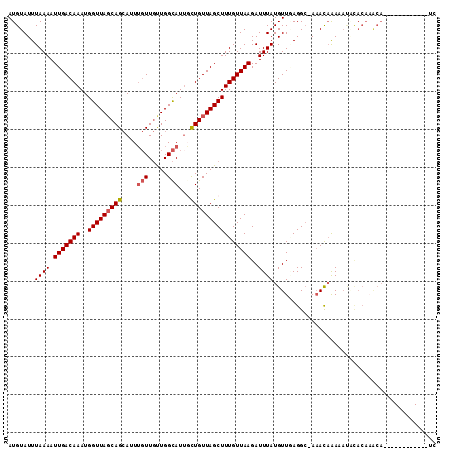
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:31 2006