| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,909,935 – 12,910,025 |
| Length | 90 |
| Max. P | 0.999432 |

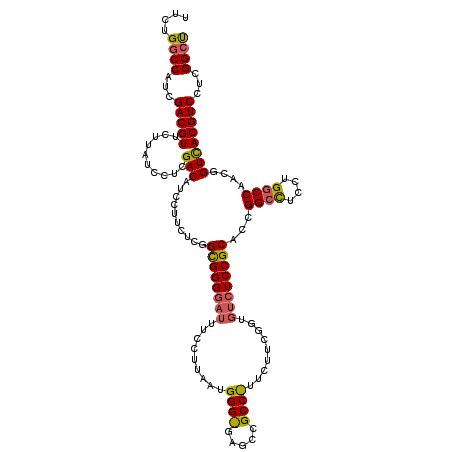
| Location | 12,909,935 – 12,910,025 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -36.90 |
| Consensus MFE | -24.05 |
| Energy contribution | -24.83 |
| Covariance contribution | 0.78 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.31 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.575256 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12909935 90 + 27905053 GGCCGAAGACGUGACCGUUGGUCAGGAGGCCGGAGCGGA------------------------------GGAAAUCCCCGCCAAGAAGGAUGUCGAGGAUAAGAACGUCAAUGGCCAGGA (((((..(((((..((.((((((....)))))).((((.------------------------------((....))))))......)).((((...))))...)))))..))))).... ( -33.10) >DroEre_CAF1 27067 120 + 1 AGCCGAGGACGUAACCGUUGGCCAGGAGGCCGGUGCGGAGACACCGAAGAAGGGCGGCUCACCCAUUAAGGAAAUCCCCACAGAGAAGGAUGUCAAGGAUAAGAACGUCGAUGGCCAGAA .((((..(((((..(((((((((....)))))..)))).((((((......(((.......))).....((......))........)).))))..........)))))..))))..... ( -38.30) >DroYak_CAF1 28120 120 + 1 AGCCCAGGACGUGACCGUUGGCCAGGAAGCCGGUGCGGAGACACCGAAGAAAGGCGGCUCGCCCAUUAAGGAGAUCCCCGCCGAGAAGGAUGUCGAGAGUAAGAACGUCGAUGGACAGAA .(.(((.(((((..((((((((......))))..)))).((((((.......(((((.((.((......)).))...))))).....)).))))..........)))))..))).).... ( -39.30) >consensus AGCCGAGGACGUGACCGUUGGCCAGGAGGCCGGUGCGGAGACACCGAAGAA_GGCGGCUC_CCCAUUAAGGAAAUCCCCGCCGAGAAGGAUGUCGAGGAUAAGAACGUCGAUGGCCAGAA .((((..(((((..(((((((((....)))))..))))................................((.((((..........)))).))..........)))))..))))..... (-24.05 = -24.83 + 0.78)

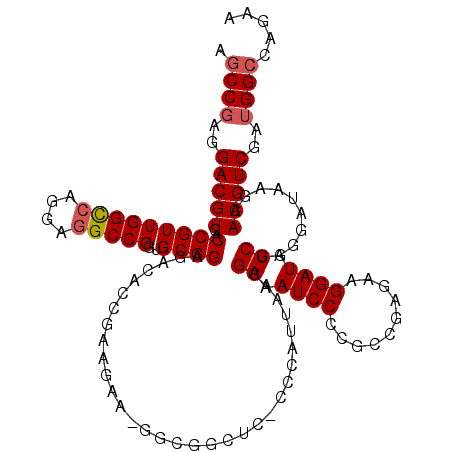
| Location | 12,909,935 – 12,910,025 |
|---|---|
| Length | 90 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 73.33 |
| Mean single sequence MFE | -36.73 |
| Consensus MFE | -28.34 |
| Energy contribution | -29.24 |
| Covariance contribution | 0.90 |
| Combinations/Pair | 1.21 |
| Mean z-score | -2.12 |
| Structure conservation index | 0.77 |
| SVM decision value | 3.60 |
| SVM RNA-class probability | 0.999432 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12909935 90 - 27905053 UCCUGGCCAUUGACGUUCUUAUCCUCGACAUCCUUCUUGGCGGGGAUUUCC------------------------------UCCGCUCCGGCCUCCUGACCAACGGUCACGUCUUCGGCC ....((((...(((((......................(((((((.....)------------------------------))))))..((((...........)))))))))...)))) ( -27.80) >DroEre_CAF1 27067 120 - 1 UUCUGGCCAUCGACGUUCUUAUCCUUGACAUCCUUCUCUGUGGGGAUUUCCUUAAUGGGUGAGCCGCCCUUCUUCGGUGUCUCCGCACCGGCCUCCUGGCCAACGGUUACGUCCUCGGCU ....((((...(((((......((..............(((((((((..((.....(((((...)))))......)).)))))))))..((((....))))...))..)))))...)))) ( -41.20) >DroYak_CAF1 28120 120 - 1 UUCUGUCCAUCGACGUUCUUACUCUCGACAUCCUUCUCGGCGGGGAUCUCCUUAAUGGGCGAGCCGCCUUUCUUCGGUGUCUCCGCACCGGCUUCCUGGCCAACGGUCACGUCCUGGGCU ....(((((..(((((..........(((..........((((((((..((.....(((((...)))))......)).))))))))...((((....))))....)))))))).))))). ( -41.20) >consensus UUCUGGCCAUCGACGUUCUUAUCCUCGACAUCCUUCUCGGCGGGGAUUUCCUUAAUGGG_GAGCCGCC_UUCUUCGGUGUCUCCGCACCGGCCUCCUGGCCAACGGUCACGUCCUCGGCU ....((((...(((((..........(((..........((((((((.........((((.....)))).........))))))))...((((....))))....))))))))...)))) (-28.34 = -29.24 + 0.90)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:42:01 2006