
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,906,975 – 12,907,158 |
| Length | 183 |
| Max. P | 0.999999 |

| Location | 12,906,975 – 12,907,078 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 72.94 |
| Mean single sequence MFE | -31.00 |
| Consensus MFE | -12.98 |
| Energy contribution | -13.10 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.14 |
| Mean z-score | -3.94 |
| Structure conservation index | 0.42 |
| SVM decision value | 1.46 |
| SVM RNA-class probability | 0.956059 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12906975 103 + 27905053 AUAACAGUGAUAAUUCCGCAUAAAAGAGGGCUUGGCUAUAAAUACAUG---------UGUGCCACACAC--UCACACACU------CGCAUCUGCAUGGCAAACCUCGCAAAAACAUUCA ......(((.......)))......((((.....(((((.......((---------((((........--.))))))..------.((....)))))))...))))............. ( -20.20) >DroEre_CAF1 24196 109 + 1 ACAAUACCGAUGAUUCCCCAUCAAAGAGGGCUUGCCUAUAAAUGCAUACUGUAUGUGUGUAC-AUACAUGUGUGCAAACA------UGCAUCUGCAUGGCAAGCCUCGCAAAAACA---- ........((((......))))...((((.((((((......((((((((((((((....))-))))).)))))))...(------(((....)))))))))))))).........---- ( -41.40) >DroYak_CAF1 25237 105 + 1 ACAAAAGUGAUGAUUU-UCAUCAAAGAGGGCUUGGCUAUAAAUACAUA---------UGUAC-AUACAUGUGUGCGCACACUAAUGUGCAUCUGCAUGGCAAACCUCGCAAAAACA---- .......(((((....-.)))))..((((.....(((((...((((((---------(((..-..))))))))).(((((....)))))......)))))...)))).........---- ( -31.40) >consensus ACAAAAGUGAUGAUUCCCCAUCAAAGAGGGCUUGGCUAUAAAUACAUA_________UGUAC_AUACAUGUGUGCACACA______UGCAUCUGCAUGGCAAACCUCGCAAAAACA____ ........((((......))))...((((..(((.((((..................(((((.........)))))...........((....))))))))).))))............. (-12.98 = -13.10 + 0.12)



| Location | 12,907,015 – 12,907,118 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 70.12 |
| Mean single sequence MFE | -25.23 |
| Consensus MFE | -13.77 |
| Energy contribution | -13.00 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.24 |
| Mean z-score | -3.56 |
| Structure conservation index | 0.55 |
| SVM decision value | 5.21 |
| SVM RNA-class probability | 0.999979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12907015 103 + 27905053 AAUACAUG---------UGUGCCACACAC--UCACACACU------CGCAUCUGCAUGGCAAACCUCGCAAAAACAUUCAUUCUCUUCAUUUCUCAUUUUUGCAGUCUUUCGCUGCAAUU ......((---------((((........--.))))))..------......(((..((....))..))).............................(((((((.....))))))).. ( -16.90) >DroEre_CAF1 24236 97 + 1 AAUGCAUACUGUAUGUGUGUAC-AUACAUGUGUGCAAACA------UGCAUCUGCAUGGCAAGCCUCGCAAAAACA----------------CUCAUUUUUGCAGUCUUUCGCUGCAGUU ..((((((((((((((....))-))))).)))))))..((------(((....)))))(((.((...(((((((..----------------....))))))).(.....)))))).... ( -32.40) >DroYak_CAF1 25276 94 + 1 AAUACAUA---------UGUAC-AUACAUGUGUGCGCACACUAAUGUGCAUCUGCAUGGCAAACCUCGCAAAAACA----------------CUUAUUUUUGCAGUCUUUCGCUGCAAUU ..((((((---------(((..-..))))))))).(((((....)))))...(((..((....))..)))......----------------.......(((((((.....))))))).. ( -26.40) >consensus AAUACAUA_________UGUAC_AUACAUGUGUGCACACA______UGCAUCUGCAUGGCAAACCUCGCAAAAACA________________CUCAUUUUUGCAGUCUUUCGCUGCAAUU .................(((((.........)))))................(((..((....))..))).............................(((((((.....))))))).. (-13.77 = -13.00 + -0.77)

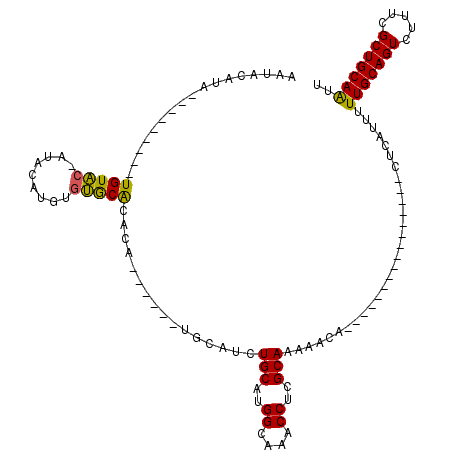

| Location | 12,907,015 – 12,907,118 |
|---|---|
| Length | 103 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 70.12 |
| Mean single sequence MFE | -32.00 |
| Consensus MFE | -20.10 |
| Energy contribution | -20.33 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.17 |
| Mean z-score | -3.91 |
| Structure conservation index | 0.63 |
| SVM decision value | 6.74 |
| SVM RNA-class probability | 0.999999 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12907015 103 - 27905053 AAUUGCAGCGAAAGACUGCAAAAAUGAGAAAUGAAGAGAAUGAAUGUUUUUGCGAGGUUUGCCAUGCAGAUGCG------AGUGUGUGA--GUGUGUGGCACA---------CAUGUAUU ..(((((((....).))))))..............((((((....))))))(((.((....)).))).((((((------.((((((.(--.....).)))))---------).)))))) ( -28.90) >DroEre_CAF1 24236 97 - 1 AACUGCAGCGAAAGACUGCAAAAAUGAG----------------UGUUUUUGCGAGGCUUGCCAUGCAGAUGCA------UGUUUGCACACAUGUAU-GUACACACAUACAGUAUGCAUU ...((((((....).))))).......(----------------(((.((((((.((....)).)))))).)))------)...((((.((.(((((-((....))))))))).)))).. ( -34.30) >DroYak_CAF1 25276 94 - 1 AAUUGCAGCGAAAGACUGCAAAAAUAAG----------------UGUUUUUGCGAGGUUUGCCAUGCAGAUGCACAUUAGUGUGCGCACACAUGUAU-GUACA---------UAUGUAUU ..(((((((....).))))))......(----------------(((.((((((.((....)).)))))).))))....(((((((.((....)).)-)))))---------)....... ( -32.80) >consensus AAUUGCAGCGAAAGACUGCAAAAAUGAG________________UGUUUUUGCGAGGUUUGCCAUGCAGAUGCA______UGUGUGCACACAUGUAU_GUACA_________UAUGUAUU ..(((((((....).)))))).......................(((.((((((.((....)).)))))).))).......((((..((....))...)))).................. (-20.10 = -20.33 + 0.23)



| Location | 12,907,044 – 12,907,158 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -22.80 |
| Consensus MFE | -17.57 |
| Energy contribution | -17.13 |
| Covariance contribution | -0.44 |
| Combinations/Pair | 1.12 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.77 |
| SVM decision value | 1.44 |
| SVM RNA-class probability | 0.953766 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12907044 114 + 27905053 ------CGCAUCUGCAUGGCAAACCUCGCAAAAACAUUCAUUCUCUUCAUUUCUCAUUUUUGCAGUCUUUCGCUGCAAUUGUGUGCAUUAUUUUUUAUGUAUUUUGCGCAGCAUCGUUCG ------((.((((((..((....))..((((((.....................(((..(((((((.....)))))))..)))(((((........))))))))))))))).)))).... ( -25.70) >DroEre_CAF1 24275 96 + 1 ------UGCAUCUGCAUGGCAAGCCUCGCAAAAACA----------------CUCAUUUUUGCAGUCUUUCGCUGCAGUUGUGUUUAUUAUUUUUUAUGUAUU-U-UGCAGCAUCGUUCG ------((((..((((((((.......))..(((((----------------(......(((((((.....)))))))..)))))).........))))))..-.-)))).......... ( -21.70) >DroYak_CAF1 25306 102 + 1 CUAAUGUGCAUCUGCAUGGCAAACCUCGCAAAAACA----------------CUUAUUUUUGCAGUCUUUCGCUGCAAUUGUGUUCAUUAUUU-UUAUGUAUUUU-UGCAACAUCGUUUG ...(((((((..((((((((.......))...((((----------------(......(((((((.....)))))))..)))))........-.))))))....-))).))))...... ( -21.00) >consensus ______UGCAUCUGCAUGGCAAACCUCGCAAAAACA________________CUCAUUUUUGCAGUCUUUCGCUGCAAUUGUGUUCAUUAUUUUUUAUGUAUUUU_UGCAGCAUCGUUCG .......((...(((..((....))..)))........................(((..(((((((.....)))))))..)))........................))........... (-17.57 = -17.13 + -0.44)

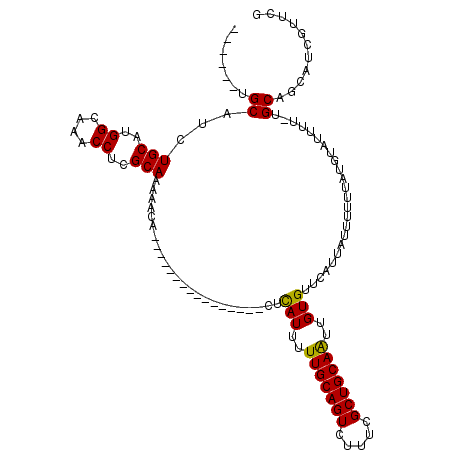

| Location | 12,907,044 – 12,907,158 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.82 |
| Mean single sequence MFE | -25.80 |
| Consensus MFE | -20.11 |
| Energy contribution | -20.00 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.99 |
| Structure conservation index | 0.78 |
| SVM decision value | 2.92 |
| SVM RNA-class probability | 0.997750 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12907044 114 - 27905053 CGAACGAUGCUGCGCAAAAUACAUAAAAAAUAAUGCACACAAUUGCAGCGAAAGACUGCAAAAAUGAGAAAUGAAGAGAAUGAAUGUUUUUGCGAGGUUUGCCAUGCAGAUGCG------ ....((...((((((((((.((((..................(((((((....).))))))...(.(....).).........))))))))))..((....))..))))...))------ ( -27.10) >DroEre_CAF1 24275 96 - 1 CGAACGAUGCUGCA-A-AAUACAUAAAAAAUAAUAAACACAACUGCAGCGAAAGACUGCAAAAAUGAG----------------UGUUUUUGCGAGGCUUGCCAUGCAGAUGCA------ ...........(((-.-..................(((((...((((((....).))))).......)----------------))))((((((.((....)).))))))))).------ ( -22.30) >DroYak_CAF1 25306 102 - 1 CAAACGAUGUUGCA-AAAAUACAUAA-AAAUAAUGAACACAAUUGCAGCGAAAGACUGCAAAAAUAAG----------------UGUUUUUGCGAGGUUUGCCAUGCAGAUGCACAUUAG .....(((((((((-(((((((....-......((....)).(((((((....).))))))......)----------------))))))))))).((((((...))))))...)))).. ( -28.00) >consensus CGAACGAUGCUGCA_AAAAUACAUAAAAAAUAAUGAACACAAUUGCAGCGAAAGACUGCAAAAAUGAG________________UGUUUUUGCGAGGUUUGCCAUGCAGAUGCA______ ..........((((............................(((((((....).))))))...........................((((((.((....)).))))))))))...... (-20.11 = -20.00 + -0.11)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:58 2006