
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,894,810 – 12,894,904 |
| Length | 94 |
| Max. P | 0.939172 |

| Location | 12,894,810 – 12,894,904 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | forward |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -40.65 |
| Consensus MFE | -26.36 |
| Energy contribution | -24.92 |
| Covariance contribution | -1.44 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 1.28 |
| SVM RNA-class probability | 0.939172 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
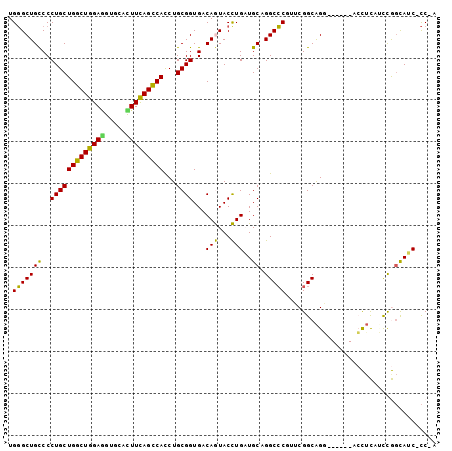
>3R_DroMel_CAF1 12894810 94 + 27905053 UGGGCUGCCCCUGCUGGCUGGAGGUUCACUUUAGUCAUCUGCGGUGACAGUACCUGAUGCUGGCCCGUUCGGCAGG------ACCUCAUUCGGCAUC-CC-A .(((.((((((((((((((((((.....))))))))....((((.(.(((((.....))))).)))))..))))))------.........)))).)-))-. ( -38.90) >DroPse_CAF1 9337 95 + 1 UGGGCUGUCCCUGCUGGCUGGAAGUGCAUUUCAGCCACCUGCGGUGACAAUACUUGCUGCUGGCCCUUACGGCAGU------AAUUUCACCAGCAGCGCC-C .((((((((.(((((((((((((.....)))))))))...)))).))))......((((((((...((((....))------)).....)))))))))))-) ( -46.10) >DroSim_CAF1 13427 94 + 1 UGGGCUGCCCCUGCUGGCUGGAGGUUCACUUUAGCCAUCUGCGGUGACAGUACCUGAUGCAGGCCCGUUCGGCAGG------ACCUCAUUCGGCAUC-CC-A .(((.((((((((((((((((((.....)))))))).(((((((((....))))....))))).......))))))------.........)))).)-))-. ( -39.30) >DroYak_CAF1 12804 94 + 1 UGGGCUGCCCCUGCUGGCUGGAGGUUCACUUUAGCCAUCUGCGGUGACAGUACCUGAUGCAGGCCCGUUCGGCAGG------ACCUCAUUCGGUAAC-CC-A .((((((.(.(((((((((((((.....)))))))))...)))).).)))((((.((((.(((.((........))------.))))))).)))).)-))-. ( -38.70) >DroAna_CAF1 10207 101 + 1 UGCGCUGCCCCUGCUGGCUGGACGUGAACUUCAGCCACCUGCGGUGACAGUACCUGAUGCAGGCGCUCUCGGCGGUCACCCUACCACACCUGCCAUC-UCUG ...((((.(.((((((((((((.(....)))))))))...)))).).))))....((((((((.((.....))(((......)))...)))).))))-.... ( -35.30) >DroPer_CAF1 10362 95 + 1 UGGGCUGUCCCUGCUGGCUGGAGGUGCAUUUCAGCCACCUGCGGUGACAAUACUUGCUGCUGGCCCUUACGGCAGU------AAUUUCACCAGCAGCGCC-C .((((((((.(((((((((((((.....)))))))))...)))).))))......((((((((...((((....))------)).....)))))))))))-) ( -45.60) >consensus UGGGCUGCCCCUGCUGGCUGGAGGUGCACUUCAGCCACCUGCGGUGACAGUACCUGAUGCAGGCCCGUUCGGCAGG______ACCUCAUCCGGCAUC_CC_A .(((((((..(((((((((((((.....)))))))))...))))...(((...)))..)).))))).................................... (-26.36 = -24.92 + -1.44)


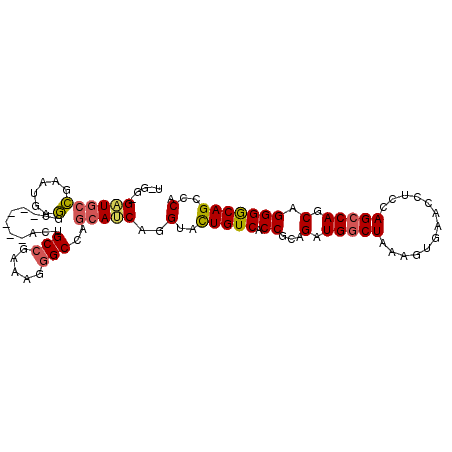
| Location | 12,894,810 – 12,894,904 |
|---|---|
| Length | 94 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 79.99 |
| Mean single sequence MFE | -40.72 |
| Consensus MFE | -21.01 |
| Energy contribution | -20.62 |
| Covariance contribution | -0.39 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.38 |
| Structure conservation index | 0.52 |
| SVM decision value | 0.55 |
| SVM RNA-class probability | 0.777789 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12894810 94 - 27905053 U-GG-GAUGCCGAAUGAGGU------CCUGCCGAACGGGCCAGCAUCAGGUACUGUCACCGCAGAUGACUAAAGUGAACCUCCAGCCAGCAGGGGCAGCCCA .-((-(.((((....(((((------((((((.....))).((((((.(((......)))...)))).))..)).).)))))...((....)))))).))). ( -30.80) >DroPse_CAF1 9337 95 - 1 G-GGCGCUGCUGGUGAAAUU------ACUGCCGUAAGGGCCAGCAGCAAGUAUUGUCACCGCAGGUGGCUGAAAUGCACUUCCAGCCAGCAGGGACAGCCCA (-((((((((((((....((------((....))))..)))))))))......((((.((...(.((((((...........)))))).).)))))))))). ( -46.20) >DroSim_CAF1 13427 94 - 1 U-GG-GAUGCCGAAUGAGGU------CCUGCCGAACGGGCCUGCAUCAGGUACUGUCACCGCAGAUGGCUAAAGUGAACCUCCAGCCAGCAGGGGCAGCCCA .-((-(.((((..(((((((------((........)))))).)))..))))(((((.((((...(((((..((.....))..))))))).)))))))))). ( -38.10) >DroYak_CAF1 12804 94 - 1 U-GG-GUUACCGAAUGAGGU------CCUGCCGAACGGGCCUGCAUCAGGUACUGUCACCGCAGAUGGCUAAAGUGAACCUCCAGCCAGCAGGGGCAGCCCA .-((-(.((((..(((((((------((........)))))).)))..))))(((((.((((...(((((..((.....))..))))))).)))))))))). ( -37.50) >DroAna_CAF1 10207 101 - 1 CAGA-GAUGGCAGGUGUGGUAGGGUGACCGCCGAGAGCGCCUGCAUCAGGUACUGUCACCGCAGGUGGCUGAAGUUCACGUCCAGCCAGCAGGGGCAGCGCA ....-((((.((((((((((.((....)))))....)))))))))))..(..(((((.((...(.((((((..(....)...)))))).).)))))))..). ( -45.50) >DroPer_CAF1 10362 95 - 1 G-GGCGCUGCUGGUGAAAUU------ACUGCCGUAAGGGCCAGCAGCAAGUAUUGUCACCGCAGGUGGCUGAAAUGCACCUCCAGCCAGCAGGGACAGCCCA (-((((((((((((....((------((....))))..)))))))))......((((.((...(.((((((...........)))))).).)))))))))). ( -46.20) >consensus U_GG_GAUGCCGAAUGAGGU______ACUGCCGAAAGGGCCAGCAUCAGGUACUGUCACCGCAGAUGGCUAAAGUGAACCUCCAGCCAGCAGGGGCAGCCCA .....((((((......)...........(((.....)))..)))))..(..(((((.((...(.(((((.............))))).).)))))))..). (-21.01 = -20.62 + -0.39)

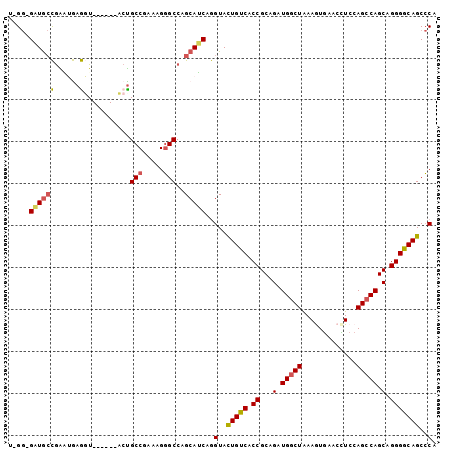
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:53 2006