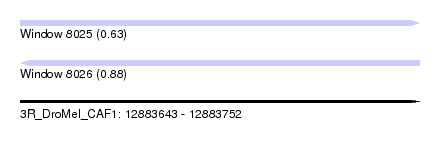
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,883,643 – 12,883,752 |
| Length | 109 |
| Max. P | 0.881414 |

| Location | 12,883,643 – 12,883,752 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 66.42 |
| Mean single sequence MFE | -30.33 |
| Consensus MFE | -11.22 |
| Energy contribution | -15.78 |
| Covariance contribution | 4.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -1.73 |
| Structure conservation index | 0.37 |
| SVM decision value | 0.19 |
| SVM RNA-class probability | 0.627660 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12883643 109 + 27905053 GUUCGAUAGCCUGCGGGAUGGACUGCUCCAGCUCAAACUCCAGCUCCUGUGGCCAGAGUUUGAGUCUGAG------UCAGGGGCAAGGCCACAACAACAACAACAGCCAUCCGUA ...............((((((.((((((..((((((((((..(((.....)))..))))))))))..)))------.))).(((...)))................))))))... ( -38.20) >DroPse_CAF1 92 85 + 1 AUUCGAUUCCCUACGCGACAGACUGCUGCGGCCCC------------UACUGCCAGAACA----------------ACAGCGGCA--GCCCCAACGCCAACAACUGCCGCCUCCC ..............(((.(((.((((((((((...------------....))).(....----------------.).))))))--).......(....)..))).)))..... ( -18.90) >DroSec_CAF1 1400 109 + 1 GUUCGAUAGCCUGCGGGAUGGACUGCUCCAGCUCAAACUCCAGCUCCUGUGGCCAGAGUUUGAGUCUGAG------UCAGGGGCAAAGCCACAACAACAACAACAGCCAUCCGUA ...............((((((.((((((..((((((((((..(((.....)))..))))))))))..)))------.))).(((...)))................))))))... ( -38.20) >DroSim_CAF1 1 92 + 1 -----------------AUGGACUGCUCCAGCUCAAACUCCAGCUCCUGUGGCCAGAGUUUGAGUCUGAG------UCAGGGGCAAAGCCACAACAACAACAACAGCCAUCCGUA -----------------(((((((((((..((((((((((..(((.....)))..))))))))))..)))------.))).(((...)))...................))))). ( -32.00) >DroYak_CAF1 1507 115 + 1 GUUCAAUGUCCUGCGCGAUGGACUGCUCCAGCUCAAACUCCAGCUCCUGUGGCCAGAGUUUGAGUCUGAGUCAGAGUCAGGGGCAAAACCACAACAACAACAACAGCCAUCCGUA ...........((((.(((((.((((((..((((((((((..(((.....)))..))))))))))..))).))).((...((......))...))...........))))))))) ( -38.10) >DroPer_CAF1 192 85 + 1 AUUCGAUUCCCUACGCGACAGACUGCUGCGGCCCU------------UACUGCCAGAACA----------------ACAGCGGCA--GCCCCAACGCCAACAACUCCCGCCUCCC ..............(((...(.((((((((((...------------....))).(....----------------.).))))))--).)....))).................. ( -16.60) >consensus GUUCGAUACCCUGCGCGAUGGACUGCUCCAGCUCAAACUCCAGCUCCUGUGGCCAGAGUUUGAGUCUGAG______UCAGGGGCAAAGCCACAACAACAACAACAGCCAUCCGUA ..................(((..((((((.((((((((((..(((.....)))..))))))))))..............))))))...)))........................ (-11.22 = -15.78 + 4.56)

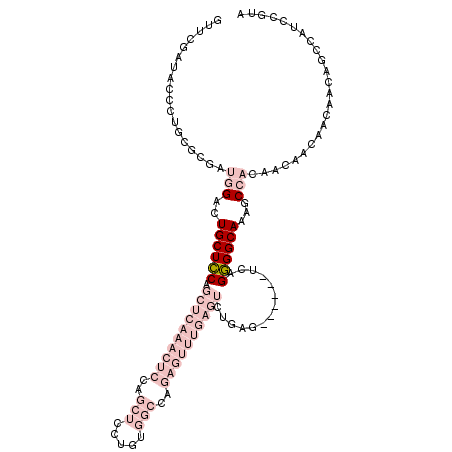

| Location | 12,883,643 – 12,883,752 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 66.42 |
| Mean single sequence MFE | -37.93 |
| Consensus MFE | -18.99 |
| Energy contribution | -23.97 |
| Covariance contribution | 4.97 |
| Combinations/Pair | 1.13 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.50 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881414 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12883643 109 - 27905053 UACGGAUGGCUGUUGUUGUUGUUGUGGCCUUGCCCCUGA------CUCAGACUCAAACUCUGGCCACAGGAGCUGGAGUUUGAGCUGGAGCAGUCCAUCCCGCAGGCUAUCGAAC ...(((((((((((......((((.(((...))).).))------)((((.(((((((((..(((....).))..)))))))))))))))))).))))))............... ( -42.70) >DroPse_CAF1 92 85 - 1 GGGAGGCGGCAGUUGUUGGCGUUGGGGC--UGCCGCUGU----------------UGUUCUGGCAGUA------------GGGGCCGCAGCAGUCUGUCGCGUAGGGAAUCGAAU .....(((((((..(....)((((.(((--(.(..((((----------------(.....)))))..------------).)))).))))...))))))).............. ( -33.00) >DroSec_CAF1 1400 109 - 1 UACGGAUGGCUGUUGUUGUUGUUGUGGCUUUGCCCCUGA------CUCAGACUCAAACUCUGGCCACAGGAGCUGGAGUUUGAGCUGGAGCAGUCCAUCCCGCAGGCUAUCGAAC ...(((((((((((......((((.(((...))).).))------)((((.(((((((((..(((....).))..)))))))))))))))))).))))))............... ( -42.70) >DroSim_CAF1 1 92 - 1 UACGGAUGGCUGUUGUUGUUGUUGUGGCUUUGCCCCUGA------CUCAGACUCAAACUCUGGCCACAGGAGCUGGAGUUUGAGCUGGAGCAGUCCAU----------------- ...(((.(((....((..(....)..))...))).(((.------.((((.(((((((((..(((....).))..)))))))))))))..))))))..----------------- ( -37.60) >DroYak_CAF1 1507 115 - 1 UACGGAUGGCUGUUGUUGUUGUUGUGGUUUUGCCCCUGACUCUGACUCAGACUCAAACUCUGGCCACAGGAGCUGGAGUUUGAGCUGGAGCAGUCCAUCGCGCAGGACAUUGAAC ..((((((((((((((((..((((.((......)).))))..))))((((.(((((((((..(((....).))..)))))))))))))))))).))))).))............. ( -42.80) >DroPer_CAF1 192 85 - 1 GGGAGGCGGGAGUUGUUGGCGUUGGGGC--UGCCGCUGU----------------UGUUCUGGCAGUA------------AGGGCCGCAGCAGUCUGUCGCGUAGGGAAUCGAAU ....((((((.(((((.(((.((...((--(((((....----------------.....))))))).------------)).))))))))..))))))................ ( -28.80) >consensus UACGGAUGGCUGUUGUUGUUGUUGUGGCUUUGCCCCUGA______CUCAGACUCAAACUCUGGCCACAGGAGCUGGAGUUUGAGCUGGAGCAGUCCAUCGCGCAGGCAAUCGAAC ....((((((((((.........(.(((...))).)...........(((.(((((((((((((.......)))))))))))))))).))))).)))))................ (-18.99 = -23.97 + 4.97)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:39 2006