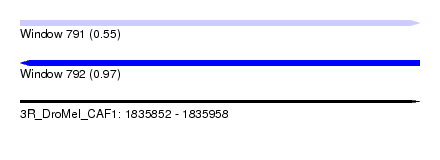
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,835,852 – 1,835,958 |
| Length | 106 |
| Max. P | 0.972890 |

| Location | 1,835,852 – 1,835,958 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -27.33 |
| Consensus MFE | -15.31 |
| Energy contribution | -15.53 |
| Covariance contribution | 0.23 |
| Combinations/Pair | 1.15 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.56 |
| SVM decision value | 0.02 |
| SVM RNA-class probability | 0.545413 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
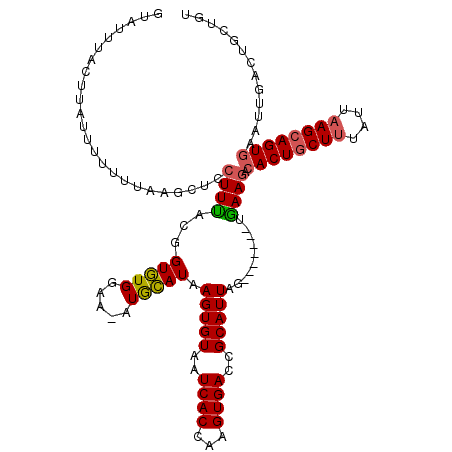
>3R_DroMel_CAF1 1835852 106 + 27905053 -UAU---GUUAUAAUUUUAUCCUCCUUUGGGGUAUGGAA-AUAUAUAAGUGUGAUCACCAAGUGACCGCAUUAGCAAGCAGUAAAGGCACUGCUUUAUCUAGCAGU---------GUUAU -..(---((((((.((((((((((....)))).))))))-.))))..((((((.((((...)))).)))))).))).........(((((((((......))))))---------))).. ( -29.60) >DroSec_CAF1 2103 113 + 1 GUAUUUACUUAUUUUUUUAAGCUCCUUUACGGUGUGGAAAAUGCAUAAGUGUAAUCACCAAGUGACCGCAUUAG-------AGAAGACACUGCUUUAUUAAGCAGUGAAUUGACUGCUGU ...................(((..(((((..((((((....((((....)))).((((...)))))))))))))-------))....((((((((....))))))))........))).. ( -27.40) >DroSim_CAF1 2015 113 + 1 GUAUUUACUUAUUUUUUUAAGCUCCUUCACGGUGUGAAAGAUGCAUAAGUGUAAUCACCAAGUGACCGCAUUAG-------UGAAAACACUACUUUAUUAAGCAGUGAAUUGACUGCUGU ((.((((((...........((((.(((((...))))).)).))...(((((..((((...))))..)))))))-------)))).))............((((((......)))))).. ( -25.00) >consensus GUAUUUACUUAUUUUUUUAAGCUCCUUUACGGUGUGGAA_AUGCAUAAGUGUAAUCACCAAGUGACCGCAUUAG_______UGAAGACACUGCUUUAUUAAGCAGUGAAUUGACUGCUGU ........................((((...(((((.....))))).(((((..((((...))))..)))))..........)))).((((((((....))))))))............. (-15.31 = -15.53 + 0.23)

| Location | 1,835,852 – 1,835,958 |
|---|---|
| Length | 106 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 75.35 |
| Mean single sequence MFE | -24.93 |
| Consensus MFE | -14.68 |
| Energy contribution | -15.13 |
| Covariance contribution | 0.45 |
| Combinations/Pair | 1.14 |
| Mean z-score | -2.25 |
| Structure conservation index | 0.59 |
| SVM decision value | 1.70 |
| SVM RNA-class probability | 0.972890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
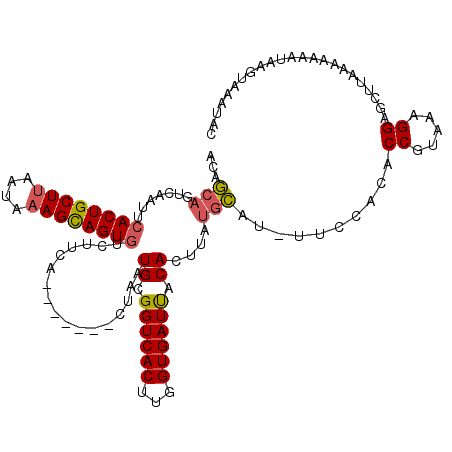
>3R_DroMel_CAF1 1835852 106 - 27905053 AUAAC---------ACUGCUAGAUAAAGCAGUGCCUUUACUGCUUGCUAAUGCGGUCACUUGGUGAUCACACUUAUAUAU-UUCCAUACCCCAAAGGAGGAUAAAAUUAUAAC---AUA- ....(---------((((((......)))))))(((((.......((....))((((((...))))))............-...........)))))................---...- ( -21.80) >DroSec_CAF1 2103 113 - 1 ACAGCAGUCAAUUCACUGCUUAAUAAAGCAGUGUCUUCU-------CUAAUGCGGUCACUUGGUGAUUACACUUAUGCAUUUUCCACACCGUAAAGGAGCUUAAAAAAAUAAGUAAAUAC ...(((.......((((((((....))))))))......-------....((..(((((...)))))..))....)))..........((.....)).(((((......)))))...... ( -23.30) >DroSim_CAF1 2015 113 - 1 ACAGCAGUCAAUUCACUGCUUAAUAAAGUAGUGUUUUCA-------CUAAUGCGGUCACUUGGUGAUUACACUUAUGCAUCUUUCACACCGUGAAGGAGCUUAAAAAAAUAAGUAAAUAC ...(((((.((..((((((((....))))))))..)).)-------))..((..(((((...)))))..)).....)).((((((((...))))))))(((((......)))))...... ( -29.70) >consensus ACAGCAGUCAAUUCACUGCUUAAUAAAGCAGUGUCUUCA_______CUAAUGCGGUCACUUGGUGAUUACACUUAUGCAU_UUCCACACCGUAAAGGAGCUUAAAAAAAUAAGUAAAUAC ...(((.......((((((((....)))))))).................((.((((((...)))))).))....)))..........((.....))....................... (-14.68 = -15.13 + 0.45)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:29 2006