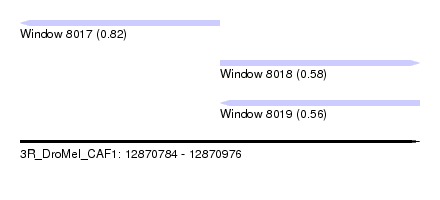
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,870,784 – 12,870,976 |
| Length | 192 |
| Max. P | 0.822775 |

| Location | 12,870,784 – 12,870,880 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 102 |
| Reading direction | reverse |
| Mean pairwise identity | 86.46 |
| Mean single sequence MFE | -40.29 |
| Consensus MFE | -31.88 |
| Energy contribution | -31.33 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.39 |
| Mean z-score | -2.71 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.69 |
| SVM RNA-class probability | 0.822775 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12870784 96 - 27905053 UUGGGUAGAGGUCGCUGGUUGCCGCC---GGGCAGUGGAAAGUUGUGCGGCAUGUGCUGCUGCAUUAGAGGAUGCUG---GGGCAAUUGCGGAAUAUUCCCG ..(((...(..((((.(((((((.((---((.((..........((((((((.....)))))))).......)))))---))))))))))))..)...))). ( -38.03) >DroSec_CAF1 1212 96 - 1 UUGGGAAGAGGUCGCUGGUUGCCGCC---GGGCAGUGGAAAGUUGGGCGGCAUGUGCUGCUGCAUUAGAGGAUGCUG---AGGCAAUUGCGGAAUAUUCCCA .((((((....((((....(((((((---.(((........))).)))))))..((((...(((((....)))))..---.))))...))))....)))))) ( -42.30) >DroSim_CAF1 1274 96 - 1 UUGGGUAGAGGUCGCUGGUUGCCGCC---GGGCAGUGGAAAGUUGGGCGGCAUGUGCUGCUGCAUUAGAGGAUGCUG---AGGCAAUUGCGGAAUAUUCCCG ..(((...(..((((....(((((((---.(((........))).)))))))..((((...(((((....)))))..---.))))...))))..)...))). ( -39.20) >DroEre_CAF1 1377 96 - 1 UUGGGUAGAGGUCGCUGGUUGCCGCC---GGGCAGUGGAAAGUUUGGCGGCAUGUGUUGCUGCAUCAGCGGAUGCUG---GGGCAAUUGCGGAAUAUUCCCG ..(((...(..((((.((((((((((---((((........)))))))(((((.(((((......))))).))))).---.)))))))))))..)...))). ( -41.50) >DroYak_CAF1 1303 96 - 1 UUGGGUAGAGGUCGCUGGUUGCCGCC---GGGCAGUGGAAAGUUUGGCAGCAUGUGCUGCUGUAUUAGCGGAUGCUG---GGGUAAUUGCGGAAUAUUCCCG ..(((...(..((((.((((((((((---((((........)))))))(((((.(((((......))))).))))).---.)))))))))))..)...))). ( -40.50) >DroAna_CAF1 1177 102 - 1 UUGGGCAGUGGACGCUGAUUGCCGCCAUUGGGCAGCGGAUAGUUCAGCGGUAGGUAAUUGGGCAUCAGCGGGUGCUGCGCAGGCAGUGGUGGCAUAUUUCCG .((.((((((..(((((((.((((((....))).((.((....)).))............))))))))))..)))))).))((.((((......)))).)). ( -40.20) >consensus UUGGGUAGAGGUCGCUGGUUGCCGCC___GGGCAGUGGAAAGUUGGGCGGCAUGUGCUGCUGCAUUAGAGGAUGCUG___AGGCAAUUGCGGAAUAUUCCCG ..(((......((((.((((((((((....)))............((((((....))))))(((((....)))))......)))))))))))......))). (-31.88 = -31.33 + -0.55)



| Location | 12,870,880 – 12,870,976 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | forward |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -26.73 |
| Consensus MFE | -17.49 |
| Energy contribution | -18.05 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.25 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.09 |
| SVM RNA-class probability | 0.580869 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12870880 96 + 27905053 UCCACAAGCUCUGCCCACGGCCCUGAACAAUUUCGCGAUGCAUCCAAGCUUCAAUGCGAUGCGCGCCGCUGGAAU---CCAUCCAGGGGCUUUUAUGCA .......((...((((.((((...(((....)))((...(((((...((......)))))))))))))(((((..---...)))))))))......)). ( -30.10) >DroVir_CAF1 1949 93 + 1 CCCACU---UUUGCCCACGGCGCUCAACAAUUUUGCCAUGCAUCCGAGCUUUAAUGCAAUGCGCGCCGUUGGCAU---GCACCCGGCGGCCUUUAUGCA .((...---..((((.(((((((.((.......(((...))).....((......))..)).))))))).)))).---((.....)))).......... ( -27.50) >DroGri_CAF1 2636 96 + 1 UCCUCA---UUUGCCCACUGCACUCAACAACUUUGCCAUGCAUCCGAGCUUCAAUGCGAUGCGUGCCGUUGGCAUCCCGCACCCCGCAACCUUUAUGCA ......---..((((.((.((((.(((.....)))....(((((...((......))))))))))).)).))))...........(((.......))). ( -21.90) >DroEre_CAF1 1473 96 + 1 UCCACACGCUUUGCCCACGGCCCUCAACAAUUUUGCGAUGCAUCCGAGCUUCAAUGCGAUGCGCGCCGCUGGGCU---GCAUCCAGCCGCGUUUAUGCA .....((((...(((((((((.(.(((.....))).)..(((((...((......)))))))..)))).))))).---((.....)).))))....... ( -28.70) >DroMoj_CAF1 1447 91 + 1 UCCACA---UCUGCCCACGGCGCUCAACAACUUUGCCAUGCAUCCCAGCUUCAAUGCGAUGCGCGCCGUUGGCUU---GCAUUCAGCGGCCUACAUG-- .((...---...(((.(((((((.(((.....)))....(((((.((.......)).)))))))))))).)))..---((.....))))........-- ( -28.60) >DroPer_CAF1 1097 93 + 1 CCCACA---UCUGCCCACAGCCCUAAACAACUUUGCAAUGCAUCCAAGUUUCAAUGCAAUGCGAGCUGCUGGCUU---GCAUCCGGCGGCCUUUAUGCA ......---..........((...........(((((.(((((..........))))).)))))((((((((...---....))))))))......)). ( -23.60) >consensus UCCACA___UCUGCCCACGGCCCUCAACAACUUUGCCAUGCAUCCGAGCUUCAAUGCGAUGCGCGCCGCUGGCAU___GCAUCCAGCGGCCUUUAUGCA ............((.....)).............((..((((((...((......)))))))).((((((((..........))))))))......)). (-17.49 = -18.05 + 0.56)


| Location | 12,870,880 – 12,870,976 |
|---|---|
| Length | 96 |
| Sequences | 6 |
| Columns | 99 |
| Reading direction | reverse |
| Mean pairwise identity | 81.21 |
| Mean single sequence MFE | -33.82 |
| Consensus MFE | -21.84 |
| Energy contribution | -21.68 |
| Covariance contribution | -0.16 |
| Combinations/Pair | 1.31 |
| Mean z-score | -1.52 |
| Structure conservation index | 0.65 |
| SVM decision value | 0.05 |
| SVM RNA-class probability | 0.557118 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12870880 96 - 27905053 UGCAUAAAAGCCCCUGGAUGG---AUUCCAGCGGCGCGCAUCGCAUUGAAGCUUGGAUGCAUCGCGAAAUUGUUCAGGGCCGUGGGCAGAGCUUGUGGA .((......))(((((((..(---.(((..((((...(((((.((........))))))).))))))).)..)))))))(((..(((...)))..))). ( -34.20) >DroVir_CAF1 1949 93 - 1 UGCAUAAAGGCCGCCGGGUGC---AUGCCAACGGCGCGCAUUGCAUUAAAGCUCGGAUGCAUGGCAAAAUUGUUGAGCGCCGUGGGCAAA---AGUGGG (((((...((...))..))))---)((((.((((((((((((((......))...)))))..((((....))))..))))))).))))..---...... ( -34.50) >DroGri_CAF1 2636 96 - 1 UGCAUAAAGGUUGCGGGGUGCGGGAUGCCAACGGCACGCAUCGCAUUGAAGCUCGGAUGCAUGGCAAAGUUGUUGAGUGCAGUGGGCAAA---UGAGGA .(((.......)))(.((((((...((((...)))))))))).)......(((((..(((((..(((.....))).))))).)))))...---...... ( -29.80) >DroEre_CAF1 1473 96 - 1 UGCAUAAACGCGGCUGGAUGC---AGCCCAGCGGCGCGCAUCGCAUUGAAGCUCGGAUGCAUCGCAAAAUUGUUGAGGGCCGUGGGCAAAGCGUGUGGA ..(((..((((.((.....))---.(((((.((((.((((((((......))...))))).(((((....)).)))).)))))))))...))))))).. ( -35.70) >DroMoj_CAF1 1447 91 - 1 --CAUGUAGGCCGCUGAAUGC---AAGCCAACGGCGCGCAUCGCAUUGAAGCUGGGAUGCAUGGCAAAGUUGUUGAGCGCCGUGGGCAGA---UGUGGA --(((((..((((((......---.)))..((((((((((((.((.......)).)))))..((((....))))..))))))).)))..)---)))).. ( -36.10) >DroPer_CAF1 1097 93 - 1 UGCAUAAAGGCCGCCGGAUGC---AAGCCAGCAGCUCGCAUUGCAUUGAAACUUGGAUGCAUUGCAAAGUUGUUUAGGGCUGUGGGCAGA---UGUGGG (((((...((...))..))))---).(((.((((((((((.((((((.(....).)))))).)))(((....))).))))))).)))...---...... ( -32.60) >consensus UGCAUAAAGGCCGCUGGAUGC___AUGCCAACGGCGCGCAUCGCAUUGAAGCUCGGAUGCAUGGCAAAAUUGUUGAGGGCCGUGGGCAAA___UGUGGA ..((((......((.....)).....(((.((((((((((((((......))...)))))...(((....)))...))))))).)))......)))).. (-21.84 = -21.68 + -0.16)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:32 2006