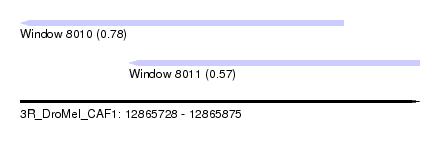
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,865,728 – 12,865,875 |
| Length | 147 |
| Max. P | 0.782056 |

| Location | 12,865,728 – 12,865,847 |
|---|---|
| Length | 119 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 80.21 |
| Mean single sequence MFE | -34.69 |
| Consensus MFE | -22.28 |
| Energy contribution | -23.53 |
| Covariance contribution | 1.25 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.85 |
| Structure conservation index | 0.64 |
| SVM decision value | 0.56 |
| SVM RNA-class probability | 0.782056 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12865728 119 - 27905053 GUUUUUUCAAAAUUAAGUGGUAAUUAUGCACCGAAUACCAUUGUAUAUGGGGUCGGAGCCUCGCCCCGAGGCCUCUGAAUAUAUCACGAUGUCCGAGGCCAUGUGUCCACGUAACCUGA ................((((....(((((..((.....(((((((((((...(((((((((((...))))).)))))).))))).))))))..))..).))))...))))......... ( -33.70) >DroSec_CAF1 35435 119 - 1 UUUUUUUCAAAAUUAAGUGGUAAAUAUGCACCGAAUACCAUUGUAUAUGGCGUCGGAGCCUCGACCCGAGGCCUCUGAAUAUAUCACGAUGUCCGAGGUCAUGUGUCCACGUAACCUGA ................((((...(((((.(((......(((((((((((...(((((((((((...))))).)))))).))))).)))))).....))))))))..))))......... ( -38.70) >DroSim_CAF1 36109 119 - 1 GUUUUUUCAAAAUUAAGUGGUAAAUAUGCACCGAAUACCAUUGUAUAUGGCGUCGGAGCCUCGACCCGAGGCCUCUGAAUAUAUCACGAUGUCCGAAGUCAUGUGUCCACGUAACCUAA ................((((...(((((.((.......(((((((((((...(((((((((((...))))).)))))).))))).))))))......)))))))..))))......... ( -34.12) >DroEre_CAF1 35234 119 - 1 UUUUCUUCCAAAUUAGGUGGUGGAUAUGCAUAGAAUACCAUUGCAUAUGGUGUCGGAGCCUUGUCCAGAGGCCUCUGAAUACAUCACGAUGUCCGGGGUCAUGUGUCCACGUAACCUGA ............((((((.((((((..((((.((...((...((((.(((((((((((((((.....)))).))))))...)))))..))))...)).))))))))))))...)))))) ( -39.80) >DroYak_CAF1 35485 118 - 1 -UUUUUUCCAAAUUAAGUGGUGAAUAUGCAUAGAAUACCAUUGCAUAUGGUGUCGGAGCCGUGCCCAGAGGCCUCUGAAUACAUCACGAUGUCCGGGGUCAUGUGUCCACGUAACCUGA -...............((((((.(((((((...........)))))))....((((((((.........)).))))))...))))))........((((.((((....)))).)))).. ( -31.00) >DroAna_CAF1 36628 118 - 1 -UAAAUUGCAAUUUCUUAUGAAAUUAUGCAUCGAAUUCCUCUGCAUCUUAUGGAUGAUCCUCAUCCCGGGGCUCCAGAAUGCGCCACAAUGUCAAUGGUCUUGACUCCGCGCAAUUUAA -((((((((((((((....))))))..((.((((...((..(((((.....(((((.....))))).((.((........)).))...))).))..))..))))....)))))))))). ( -30.80) >consensus _UUUUUUCAAAAUUAAGUGGUAAAUAUGCACCGAAUACCAUUGCAUAUGGUGUCGGAGCCUCGACCCGAGGCCUCUGAAUACAUCACGAUGUCCGAGGUCAUGUGUCCACGUAACCUGA ................((((((.(((((((...........)))))))....((((((((((.....)))).))))))...))))))........((((.((((....)))).)))).. (-22.28 = -23.53 + 1.25)


| Location | 12,865,768 – 12,865,875 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | reverse |
| Mean pairwise identity | 76.43 |
| Mean single sequence MFE | -28.23 |
| Consensus MFE | -17.00 |
| Energy contribution | -17.29 |
| Covariance contribution | 0.29 |
| Combinations/Pair | 1.50 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.573943 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12865768 107 - 27905053 CGAGAUCUU-CAAAACACGCUUUGAUGAG-GUUUUUUCAAAAUUAAGUGGUAAUUAUGCACCGAAUACCAUUGUAUAUGGGGUCGGAGCCUCGCCCCGAGGCCUCUGAA ..(((((((-((((......)))))..))-))))(..((......(((((((.((.......)).))))))).....))..)(((((((((((...))))).)))))). ( -26.90) >DroSec_CAF1 35475 107 - 1 CGAGAUCUU-AAAAUCACGCUUUGAGGAG-UUUUUUUCAAAAUUAAGUGGUAAAUAUGCACCGAAUACCAUUGUAUAUGGCGUCGGAGCCUCGACCCGAGGCCUCUGAA .(((.....-...(((((..((((((((.-...)))))))).....))))).........((((...((((.....))))..)))).((((((...))))))))).... ( -28.10) >DroSim_CAF1 36149 107 - 1 CGAGAUCUU-AAAAUCACGCUUUGAGGAG-GUUUUUUCAAAAUUAAGUGGUAAAUAUGCACCGAAUACCAUUGUAUAUGGCGUCGGAGCCUCGACCCGAGGCCUCUGAA .(((.....-...(((((..((((((((.-...)))))))).....))))).........((((...((((.....))))..)))).((((((...))))))))).... ( -28.10) >DroEre_CAF1 35274 109 - 1 CGAGAGCUUCAAAAUCGCAUUUUUAAGAAGUUUUCUUCCAAAUUAGGUGGUGGAUAUGCAUAGAAUACCAUUGCAUAUGGUGUCGGAGCCUUGUCCAGAGGCCUCUGAA .(((.(((((...(((((........((((....))))........)))))(((((.((...((.((((((.....))))))))...))..))))).)))))))).... ( -33.09) >DroYak_CAF1 35525 107 - 1 CGAGAGCUUCAAAAAUACGCUUUUAAGAG--UUUUUUCCAAAUUAAGUGGUGAAUAUGCAUAGAAUACCAUUGCAUAUGGUGUCGGAGCCGUGCCCAGAGGCCUCUGAA .(((.(((((.......(((((....(((--....)))......)))))....(((((((...........)))))))((((.((....))))))..)))))))).... ( -25.40) >DroAna_CAF1 36668 107 - 1 CUUUCGCUUCAAAACUGCAAUUGCAAGGA--UAAAUUGCAAUUUCUUAUGAAAUUAUGCAUCGAAUUCCUCUGCAUCUUAUGGAUGAUCCUCAUCCCGGGGCUCCAGAA .....(((((...........((((((((--.....(((((((((....)))))..))))......)))).))))......(((((.....))))).)))))....... ( -27.80) >consensus CGAGAGCUU_AAAAUCACGCUUUGAAGAG__UUUUUUCAAAAUUAAGUGGUAAAUAUGCACCGAAUACCAUUGCAUAUGGUGUCGGAGCCUCGACCCGAGGCCUCUGAA .................(((((((((((.....))))))))....(((((((.............))))))).......)))((((((((((.....)))).)))))). (-17.00 = -17.29 + 0.29)
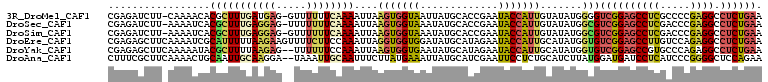
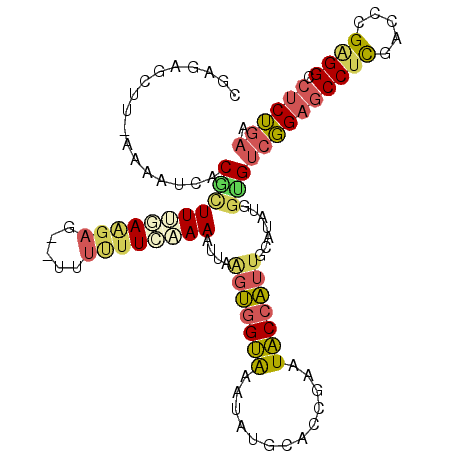
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:24 2006