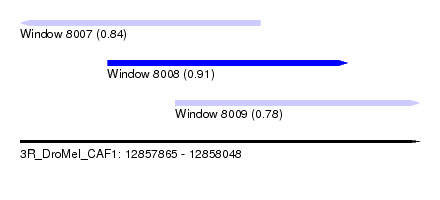
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,857,865 – 12,858,048 |
| Length | 183 |
| Max. P | 0.914696 |

| Location | 12,857,865 – 12,857,975 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 77.16 |
| Mean single sequence MFE | -32.94 |
| Consensus MFE | -14.88 |
| Energy contribution | -17.19 |
| Covariance contribution | 2.31 |
| Combinations/Pair | 1.30 |
| Mean z-score | -2.31 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.842610 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12857865 110 - 27905053 GUCCAGGUAGUCC-UGGUGGACCAAUUGGUCCCGGAAAUCCAGUUUUACCCGGCCUACCCCUUUGACCACUUUCUCCACGGUCACCACGAUCCCCCUUUUCUCCUGGCGCU ((((((((((.((-.((((((((....))))).((....))......))).)).)))))....(((((...........)))))....................))).)). ( -31.40) >DroPse_CAF1 29140 92 - 1 GACCAGU----------UGGUCCAAUUGGUCCUGCUUUUCCUCUAUUUCCUUUCCUGCCACGUAGACCACUGUCUCC---------GCGUUCACCCUUUUCACCAGAAACG (((((((----------((...)))))))))............................((((((((....))))..---------)))).......((((....)))).. ( -15.50) >DroSec_CAF1 27651 110 - 1 GGCCAGGUAGUCC-UGGUGGCCCAAUGGGUCCCGAAAAUCCAGUAUAACCCGGCCUACCCCUUUUGCCACUUUCUCCACGGUCACCACGAUCCCCCUUUUCACCUGGCGCU .(((((((.(((.-((((((((....((((.................))))(((...........)))...........)))))))).)))..........)))))))... ( -33.83) >DroSim_CAF1 28213 111 - 1 AGCCAGGUAGUCGAUGGUGGACCAAUGGGUCCCGGAAAUCCGGUAUAACCCGGCCUACCCCUUGGGCCACUUUCUCCACGGUCACCACGAUCCCCCUUUUCACCUGGCGCU .(((((((.((((.(((((.(((...((((.((((....))))....))))((((((.....))))))...........))))))))))))..........)))))))... ( -48.80) >DroEre_CAF1 27381 110 - 1 GGCCAGGUAGUCC-UGGUGGGCCAAUGGGCCCCGGAAACCCAGAAUAACCCGGCCUUCCUCUUUUUCCGCUUUCUCCGCGGUCACCAUGAUCUCCUUUUUCACCUGGCGCU .(((((((((((.-(((((((((....))))).((((..((.(......).))..)))).......((((.......))))..)))).)).))........)))))))... ( -37.90) >DroYak_CAF1 27620 110 - 1 GCCCAGGUUGUCC-AGGUGGCCCAAUGGGCCCCGGAAAUCCAGUAUAACCCGGCCUUCCUCUUUUUCCACUUUCUCCACGGUCACCACGAUCCCCCUUUUCCCCUGGCGCU (((((((......-.((.(((((...)))))))((((..((.(......).))..))))....................((((.....))))..........))))).)). ( -30.20) >consensus GGCCAGGUAGUCC_UGGUGGACCAAUGGGUCCCGGAAAUCCAGUAUAACCCGGCCUACCCCUUUGACCACUUUCUCCACGGUCACCACGAUCCCCCUUUUCACCUGGCGCU ..(((((........((((((.....((((.((((..............))))...)))).....))))))........((((.....))))..........))))).... (-14.88 = -17.19 + 2.31)


| Location | 12,857,905 – 12,858,015 |
|---|---|
| Length | 110 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 74.32 |
| Mean single sequence MFE | -39.02 |
| Consensus MFE | -20.20 |
| Energy contribution | -21.70 |
| Covariance contribution | 1.50 |
| Combinations/Pair | 1.36 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.52 |
| SVM decision value | 1.10 |
| SVM RNA-class probability | 0.914696 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12857905 110 + 27905053 AGUGGUCAAAGGGGUAGGCCGGGUAAAACUGGAUUUCCGGGACCAAUUGGUCCACCA-GGACUACCUGGACCACCAGGCCGCCCUGGCAAAUCUUCAUCGCAUGAACAGAA .(((((....(((((..((((((.....((((.......(((((....))))).(((-((....)))))....)))).....))))))..))))).))))).......... ( -42.10) >DroPse_CAF1 29171 100 + 1 AGUGGUCUACGUGGCAGGAAAGGAAAUAGAGGAAAAGCAGGACCAAUUGGACCA----------ACUGGUCCGCAAGGACUUACUAGAAAAUAUUCAACGCAUCU-AAGAA (((((((((..((((..(...................)..).)))..)))))).----------)))(((((....)))))........................-..... ( -20.81) >DroSec_CAF1 27691 110 + 1 AGUGGCAAAAGGGGUAGGCCGGGUUAUACUGGAUUUUCGGGACCCAUUGGGCCACCA-GGACUACCUGGCCCACCGGGCCGCCCUGGAAAAUCUCCAUCUCAUGGACAGAA .(((((......(((.((((((((....((((.......((.(((...))))).)))-)....)))))))).)))..))))).((((.....)(((((...)))))))).. ( -41.00) >DroSim_CAF1 28253 111 + 1 AGUGGCCCAAGGGGUAGGCCGGGUUAUACCGGAUUUCCGGGACCCAUUGGUCCACCAUCGACUACCUGGCUCACCGGGCCGCCCUGGAAAAUCUCCAUCGCAUGGACAGAA .(((((((....(((..(((((((.....(((....)))(((((....)))))..........)))))))..)))))))))).((((.....)(((((...)))))))).. ( -46.90) >DroEre_CAF1 27421 101 + 1 AGCGGAAAAAGAGGAAGGCCGGGUUAUUCUGGGUUUCCGGGGCCCAUUGGCCCACCA-GGACUACCUGGCCCACCGGGCCGCCCUGGAACAUCU---------UAUCAGAA .((((.......((..((((((((..((((((.......(((((....))))).)))-)))..))))))))..))...)))).((((.......---------..)))).. ( -43.30) >DroYak_CAF1 27660 110 + 1 AGUGGAAAAAGAGGAAGGCCGGGUUAUACUGGAUUUCCGGGGCCCAUUGGGCCACCU-GGACAACCUGGGCCAAAGGGACGCCCUGGAAAAUCUCCAUCGCAUCAACAGAA .((.((....(((((...((((......)))).((((((((((((....((((....-((....))..))))....))..))))))))))...))).))...)).)).... ( -40.00) >consensus AGUGGAAAAAGGGGUAGGCCGGGUUAUACUGGAUUUCCGGGACCCAUUGGGCCACCA_GGACUACCUGGCCCACCGGGCCGCCCUGGAAAAUCUCCAUCGCAUGAACAGAA ...........(((..((((((((....((((....)))).))))..(((((((............)))))))...))))..))).......................... (-20.20 = -21.70 + 1.50)

| Location | 12,857,936 – 12,858,048 |
|---|---|
| Length | 112 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 75.56 |
| Mean single sequence MFE | -30.58 |
| Consensus MFE | -13.80 |
| Energy contribution | -14.83 |
| Covariance contribution | 1.03 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.08 |
| Structure conservation index | 0.45 |
| SVM decision value | 0.57 |
| SVM RNA-class probability | 0.783811 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12857936 112 + 27905053 GAUUUCCGGGACCAAUUGGUCCACCA-GGACUACCUGGACCACCAGGCCGCCCUGGCAAAUCUUCAUCGCAUGAACAGAAACAAGGUACAACC-AUUAUUUACUUGCCAGCAAU ....(((.(((((....)))))....-)))...(((((....))))).....(((((((.(((((((...))))..))).....((.....))-.........))))))).... ( -32.00) >DroSec_CAF1 27722 112 + 1 GAUUUUCGGGACCCAUUGGGCCACCA-GGACUACCUGGCCCACCGGGCCGCCCUGGAAAAUCUCCAUCUCAUGGACAGAAACAAGGUUCAACC-AUUAUUUACUUGCCAGCAAU ((((((((((.(((..(((((((...-((....)))))))))..)))...)))..)))))))(((((...))))).........((.....))-.................... ( -33.40) >DroSim_CAF1 28284 113 + 1 GAUUUCCGGGACCCAUUGGUCCACCAUCGACUACCUGGCUCACCGGGCCGCCCUGGAAAAUCUCCAUCGCAUGGACAGAAACAAGGUCCAACC-AUUAUUUACUUGCCAUCAAU (((...(((((.(((.(((((.......)))))..))).)).)))(((.((..((((.....))))..)).(((((.........)))))...-...........))))))... ( -29.40) >DroEre_CAF1 27452 103 + 1 GGUUUCCGGGGCCCAUUGGCCCACCA-GGACUACCUGGCCCACCGGGCCGCCCUGGAACAUCU---------UAUCAGAAACCAGGUCCAACA-AUUAUUUACUUGCCAGCAAU ((((((.((((((....))))).(((-((.(..(((((....)))))..).))))).......---------...).)))))).(((......-...........)))...... ( -35.03) >DroYak_CAF1 27691 112 + 1 GAUUUCCGGGGCCCAUUGGGCCACCU-GGACAACCUGGGCCAAAGGGACGCCCUGGAAAAUCUCCAUCGCAUCAACAGAAACAAGGCCCAACC-AUUAUUUACUUGCCAGCAAU ((((((((((((((....((((....-((....))..))))....))..))))))).)))))......((.((....)).....(((......-...........))).))... ( -33.83) >DroAna_CAF1 28485 94 + 1 GUUUCCCCGGACCCAUAGGGCCUCCU-G------------------GGCCCCCUGGAAAAUCCGUAAUUCAUC-ACAAAAGCGAACCCCCACAAAUAAUUUAUUUGCCUGCACA .......((((.(((..(((((....-.------------------)))))..)))....)))).........-......(((......).((((((...))))))...))... ( -19.80) >consensus GAUUUCCGGGACCCAUUGGGCCACCA_GGACUACCUGGCCCACCGGGCCGCCCUGGAAAAUCUCCAUCGCAUGAACAGAAACAAGGUCCAACC_AUUAUUUACUUGCCAGCAAU ((((((((((.((...(((....))).))....(((((....)))))....))))).)))))...................((((.................))))........ (-13.80 = -14.83 + 1.03)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:23 2006