
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,855,492 – 12,855,676 |
| Length | 184 |
| Max. P | 0.977620 |

| Location | 12,855,492 – 12,855,612 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -35.67 |
| Consensus MFE | -31.45 |
| Energy contribution | -30.90 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.11 |
| Mean z-score | -2.14 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.51 |
| SVM RNA-class probability | 0.765651 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12855492 120 + 27905053 AAGGGCGGACACCACCAUCACUAUUAUCCACCAGGACACGAUAAGGGAGGCUACUAUCCGUAUCCACCCCACGGUGGCCAUGGAGGCCAUCACCAUCCAGGCCACAAUUAUCCAGGCCCC ..((((((......))....((....(((....)))...((((((((.((.(((.....))).)).)))....((((((.((((((......)).))))))))))..))))).)))))). ( -41.70) >DroSim_CAF1 25842 120 + 1 AAGGGCGGACACCACCAUCACUAUUACCCACCAGGACACGAUAAGGGAGGCCACUAUCCGUAUCCACCCCACGGCGGCCAUGGAGGCCAUCACCAUCCAGGUCAUAACUAUCCAGGACCC ..((..((..................(((....(....).....))).((((....(((((..((.((....)).))..)))))))))....))..)).((((............)))). ( -29.60) >DroYak_CAF1 25161 120 + 1 AAGGGCGGACACCACCAUCACUAUUAUCCUCCAGGACACGAUAAGGGAGGCCACUACCCGUAUCCACCCCACGGCGGCCAUGGAGACCAUCAUCAUCCAGGCCAUAAUUAUCCAGGCCCC ..(((((((.....((..........(((....)))...((((.(((((....)).))).))))........)).((((.((((((......)).)))))))).......)))..)))). ( -35.70) >consensus AAGGGCGGACACCACCAUCACUAUUAUCCACCAGGACACGAUAAGGGAGGCCACUAUCCGUAUCCACCCCACGGCGGCCAUGGAGGCCAUCACCAUCCAGGCCAUAAUUAUCCAGGCCCC ..((((((......)).................(((...((((.(((((....)).))).))))...........((((.((((((......)).)))))))).......)))..)))). (-31.45 = -30.90 + -0.55)

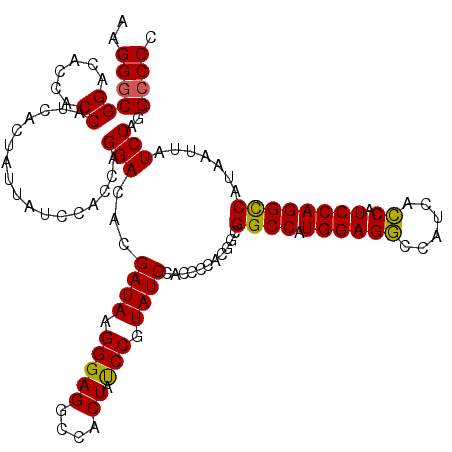
| Location | 12,855,492 – 12,855,612 |
|---|---|
| Length | 120 |
| Sequences | 3 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 93.89 |
| Mean single sequence MFE | -49.27 |
| Consensus MFE | -45.80 |
| Energy contribution | -45.70 |
| Covariance contribution | -0.10 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.82 |
| Structure conservation index | 0.93 |
| SVM decision value | 0.43 |
| SVM RNA-class probability | 0.734762 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12855492 120 - 27905053 GGGGCCUGGAUAAUUGUGGCCUGGAUGGUGAUGGCCUCCAUGGCCACCGUGGGGUGGAUACGGAUAGUAGCCUCCCUUAUCGUGUCCUGGUGGAUAAUAGUGAUGGUGGUGUCCGCCCUU ((((((.(((((...((((((((((.(((....))))))).))))))((.((((.((.(((.....))).)).))))...))))))).)))(((((.((....))....))))).))).. ( -52.60) >DroSim_CAF1 25842 120 - 1 GGGUCCUGGAUAGUUAUGACCUGGAUGGUGAUGGCCUCCAUGGCCGCCGUGGGGUGGAUACGGAUAGUGGCCUCCCUUAUCGUGUCCUGGUGGGUAAUAGUGAUGGUGGUGUCCGCCCUU .((((.((((..(((((.(((.....))).))))).)))).)))).....(((((((((((.....)).(((.((.((((..(..((....))..)...)))).)).)))))))))))). ( -46.20) >DroYak_CAF1 25161 120 - 1 GGGGCCUGGAUAAUUAUGGCCUGGAUGAUGAUGGUCUCCAUGGCCGCCGUGGGGUGGAUACGGGUAGUGGCCUCCCUUAUCGUGUCCUGGAGGAUAAUAGUGAUGGUGGUGUCCGCCCUU ((((((((((.......((((...........)))))))).)))).))..(((((((((((((((....))))((.((((..(((((....)))))...)))).))..))))))))))). ( -49.01) >consensus GGGGCCUGGAUAAUUAUGGCCUGGAUGGUGAUGGCCUCCAUGGCCGCCGUGGGGUGGAUACGGAUAGUGGCCUCCCUUAUCGUGUCCUGGUGGAUAAUAGUGAUGGUGGUGUCCGCCCUU ((((((((((..(((((.(((.....))).))))).)))).)))).))..(((((((((((.....)))(((.((.((((..(((((....)))))...)))).)).))).)))))))). (-45.80 = -45.70 + -0.10)


| Location | 12,855,532 – 12,855,651 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -35.27 |
| Consensus MFE | -31.85 |
| Energy contribution | -30.97 |
| Covariance contribution | -0.88 |
| Combinations/Pair | 1.11 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.80 |
| SVM RNA-class probability | 0.977620 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12855532 119 + 27905053 AUAAGGGAGGCUACUAUCCGUAUCCACCCCACGGUGGCCAUGGAGGCCAUCACCAUCCAGGCCACAAUUAUCCAGGCCCCUAUCCGCCACCAUAUCCACCUCACCAUCCACAUGAUGGC ....(((.((.(((.....))).)).)))...(((((..((((.((((...........))))...........(((........))).))))..)))))...(((((.....))))). ( -38.40) >DroSim_CAF1 25882 119 + 1 AUAAGGGAGGCCACUAUCCGUAUCCACCCCACGGCGGCCAUGGAGGCCAUCACCAUCCAGGUCAUAACUAUCCAGGACCCUAUCCGCCACCCUAUCCACCUCACCAUCCACAUGAUGGC ...((((.(((......((((.........)))).((((.((((((......)).))))))))...........(((.....)))))).))))..........(((((.....))))). ( -33.70) >DroYak_CAF1 25201 119 + 1 AUAAGGGAGGCCACUACCCGUAUCCACCCCACGGCGGCCAUGGAGACCAUCAUCAUCCAGGCCAUAAUUAUCCAGGCCCCUAUCCGCCACCUUAUCCACCUCACCAUCCACAUGAUGGC ....(((.((((.....((((.........)))).((((.((((((......)).))))))))...........)))))))......................(((((.....))))). ( -33.70) >consensus AUAAGGGAGGCCACUAUCCGUAUCCACCCCACGGCGGCCAUGGAGGCCAUCACCAUCCAGGCCAUAAUUAUCCAGGCCCCUAUCCGCCACCAUAUCCACCUCACCAUCCACAUGAUGGC ....(((.((..((.....))..)).)))...(((((((.((((((......)).))))))))...........((.......)))))...............(((((.....))))). (-31.85 = -30.97 + -0.88)


| Location | 12,855,532 – 12,855,651 |
|---|---|
| Length | 119 |
| Sequences | 3 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 94.12 |
| Mean single sequence MFE | -45.73 |
| Consensus MFE | -41.72 |
| Energy contribution | -41.50 |
| Covariance contribution | -0.22 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.83 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.682411 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12855532 119 - 27905053 GCCAUCAUGUGGAUGGUGAGGUGGAUAUGGUGGCGGAUAGGGGCCUGGAUAAUUGUGGCCUGGAUGGUGAUGGCCUCCAUGGCCACCGUGGGGUGGAUACGGAUAGUAGCCUCCCUUAU ((((((.....))))))..(((((.(((((.(((.(((..(((((...........)))))..)).....).))).))))).)))))..((((.((.(((.....))).)).))))... ( -48.20) >DroSim_CAF1 25882 119 - 1 GCCAUCAUGUGGAUGGUGAGGUGGAUAGGGUGGCGGAUAGGGUCCUGGAUAGUUAUGACCUGGAUGGUGAUGGCCUCCAUGGCCGCCGUGGGGUGGAUACGGAUAGUGGCCUCCCUUAU ((((((.....))))))........((.((((((((((...))))((((..(((((.(((.....))).))))).))))..)))))).))(((.((.(((.....))).)).))).... ( -45.60) >DroYak_CAF1 25201 119 - 1 GCCAUCAUGUGGAUGGUGAGGUGGAUAAGGUGGCGGAUAGGGGCCUGGAUAAUUAUGGCCUGGAUGAUGAUGGUCUCCAUGGCCGCCGUGGGGUGGAUACGGGUAGUGGCCUCCCUUAU ((((((.....))))))...........((((((......(((((...........)))))(((.(((....))))))...))))))..((((.((.(((.....))).)).))))... ( -43.40) >consensus GCCAUCAUGUGGAUGGUGAGGUGGAUAAGGUGGCGGAUAGGGGCCUGGAUAAUUAUGGCCUGGAUGGUGAUGGCCUCCAUGGCCGCCGUGGGGUGGAUACGGAUAGUGGCCUCCCUUAU ((((((.....))))))...........((((((......(((((...........)))))(((.(((....))))))...))))))..((((.((.(((.....))).)).))))... (-41.72 = -41.50 + -0.22)


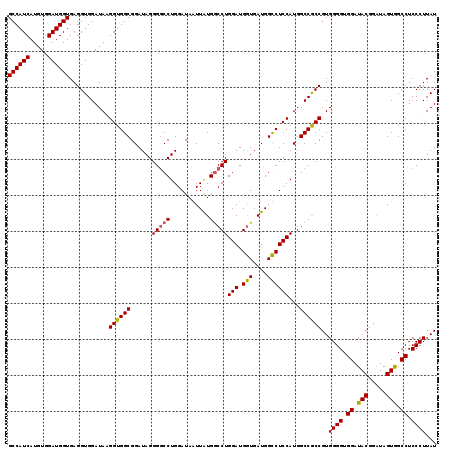
| Location | 12,855,572 – 12,855,676 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | forward |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -26.51 |
| Consensus MFE | -21.13 |
| Energy contribution | -21.68 |
| Covariance contribution | 0.56 |
| Combinations/Pair | 1.05 |
| Mean z-score | -3.07 |
| Structure conservation index | 0.80 |
| SVM decision value | 0.79 |
| SVM RNA-class probability | 0.851249 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12855572 104 + 27905053 UGGAGGCCAUCACCAUCCAGGCCACAAUUAUCCAGGCCCCUAUCCGCCACCAUAUCCACCUCACCAUCCACAUGAUGGCGGAGACAAGGGUUAUCACCAUUAUC (((.((((...........))))...........(((((...(((((((.(((..................))).))))))).....)))))....)))..... ( -28.67) >DroSim_CAF1 25922 104 + 1 UGGAGGCCAUCACCAUCCAGGUCAUAACUAUCCAGGACCCUAUCCGCCACCCUAUCCACCUCACCAUCCACAUGAUGGCGGUGACAAGGGACAUCACCAUCAUC (((.(((....(((.....)))............(((.....)))))).((((...((((...(((((.....))))).))))...))))......)))..... ( -27.60) >DroYak_CAF1 25241 104 + 1 UGGAGACCAUCAUCAUCCAGGCCAUAAUUAUCCAGGCCCCUAUCCGCCACCUUAUCCACCUCACCAUCCACAUGAUGGCGGAGACAAGGGACAUCACCAUCAUC ((((((......)).))))((((...........))))(((.(((((((.(......................).)))))))....)))............... ( -23.25) >consensus UGGAGGCCAUCACCAUCCAGGCCAUAAUUAUCCAGGCCCCUAUCCGCCACCAUAUCCACCUCACCAUCCACAUGAUGGCGGAGACAAGGGACAUCACCAUCAUC (((.((((...........))))..............((((.(((((((.(......................).)))))))....))))......)))..... (-21.13 = -21.68 + 0.56)


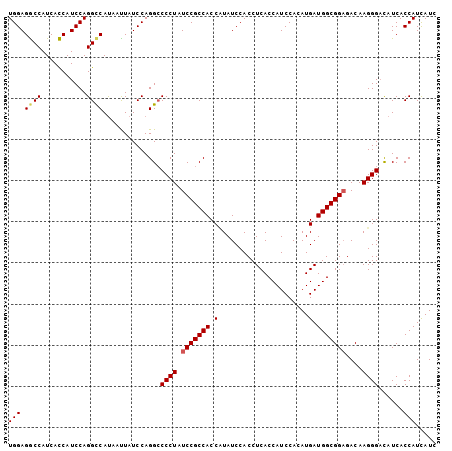
| Location | 12,855,572 – 12,855,676 |
|---|---|
| Length | 104 |
| Sequences | 3 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 92.63 |
| Mean single sequence MFE | -38.87 |
| Consensus MFE | -30.61 |
| Energy contribution | -30.07 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.17 |
| Mean z-score | -2.46 |
| Structure conservation index | 0.79 |
| SVM decision value | 0.25 |
| SVM RNA-class probability | 0.655068 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12855572 104 - 27905053 GAUAAUGGUGAUAACCCUUGUCUCCGCCAUCAUGUGGAUGGUGAGGUGGAUAUGGUGGCGGAUAGGGGCCUGGAUAAUUGUGGCCUGGAUGGUGAUGGCCUCCA (((((.(((....))).)))))(((((((((((((..((......))..)))))))))))))...(((((...........)))))(((.(((....)))))). ( -41.80) >DroSim_CAF1 25922 104 - 1 GAUGAUGGUGAUGUCCCUUGUCACCGCCAUCAUGUGGAUGGUGAGGUGGAUAGGGUGGCGGAUAGGGUCCUGGAUAGUUAUGACCUGGAUGGUGAUGGCCUCCA ............(((((((((((((((((((.....))))))..))).))))))).)))((((...)))).(((..(((((.(((.....))).))))).))). ( -37.60) >DroYak_CAF1 25241 104 - 1 GAUGAUGGUGAUGUCCCUUGUCUCCGCCAUCAUGUGGAUGGUGAGGUGGAUAAGGUGGCGGAUAGGGGCCUGGAUAAUUAUGGCCUGGAUGAUGAUGGUCUCCA (((.((.((.(((((((((((((((((((((.....))))))..)).)))))))).)))......(((((...........)))))..)).)).)).))).... ( -37.20) >consensus GAUGAUGGUGAUGUCCCUUGUCUCCGCCAUCAUGUGGAUGGUGAGGUGGAUAAGGUGGCGGAUAGGGGCCUGGAUAAUUAUGGCCUGGAUGGUGAUGGCCUCCA ......(((.(((..(((((((.((((((((.....))))))).)...))))))).(((........)))........))).))).(((.(((....)))))). (-30.61 = -30.07 + -0.55)

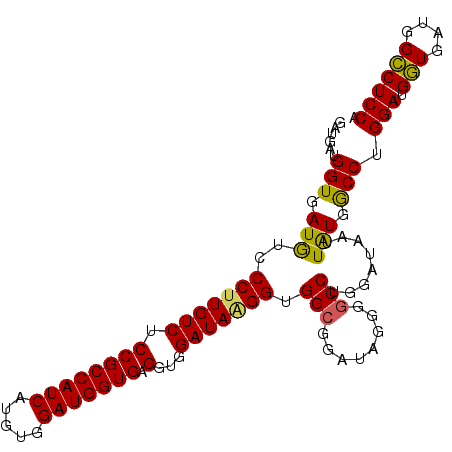

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:41:19 2006