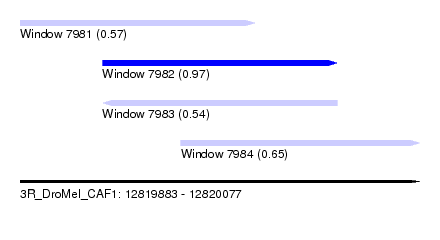
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,819,883 – 12,820,077 |
| Length | 194 |
| Max. P | 0.973890 |

| Location | 12,819,883 – 12,819,997 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 95.91 |
| Mean single sequence MFE | -32.63 |
| Consensus MFE | -28.81 |
| Energy contribution | -28.70 |
| Covariance contribution | -0.11 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.87 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.574780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12819883 114 + 27905053 UUUAACCGUCCGCUGGAUUAUCAGCGCUGCCCUCGCUGCCCUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAAAUAUGCAUAUGGUUAUUGCCAUGUACGUAUAUAAA ..........(((((......))))).(((...(((((...(((((((...)))))))....)))))....))).....(((((.(((((((....))))))).)))))..... ( -31.90) >DroSim_CAF1 197496 114 + 1 UUUAACCGUCCGCUGGAUUAUCAGCGCUGCCCUCGCUGCCCUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAAAUAUGCACAUGGUUAUUUCCAUGUACGUAUAUAAA ..........(((((......))))).(((...(((((...(((((((...)))))))....)))))....))).....(((((.((((((......)))))).)))))..... ( -32.20) >DroYak_CAF1 195750 113 + 1 UUUAACCGUCCGCUGGAUUAUCAGCGCUGCCCUCGGUGCCCUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAUAUAUACAUAUGG-UAUUGCCAUGUGCGUAUAUAAA ..........(((((........((((((....))))))..(((((((...)))))))....))))).........((((((..(((((((-.....)))))))..)))))).. ( -33.80) >consensus UUUAACCGUCCGCUGGAUUAUCAGCGCUGCCCUCGCUGCCCUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAAAUAUGCAUAUGGUUAUUGCCAUGUACGUAUAUAAA ..........(((((......))))).(((...(((((...(((((((...)))))))....)))))....))).....((((((((((((......))))))..))))))... (-28.81 = -28.70 + -0.11)

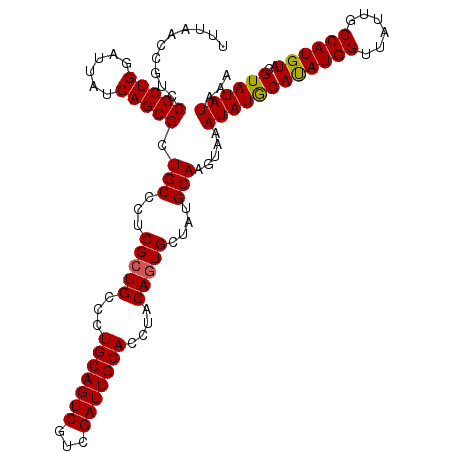

| Location | 12,819,923 – 12,820,037 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -34.50 |
| Consensus MFE | -31.87 |
| Energy contribution | -31.10 |
| Covariance contribution | -0.77 |
| Combinations/Pair | 1.13 |
| Mean z-score | -2.78 |
| Structure conservation index | 0.92 |
| SVM decision value | 1.72 |
| SVM RNA-class probability | 0.973890 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12819923 114 + 27905053 CUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAAAUAUGCAUAUGGUUAUUGCCAUGUACGUAUAUAAAUUUGCAAUUGUAUGACGAUGGACACGCACAAACCUAGUGA .(((.(((((((...((((.....))))..(((((((..(((((((((((((....)))))))..))))))..)))))))......)))))))(....))))............ ( -33.10) >DroSim_CAF1 197536 114 + 1 CUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAAAUAUGCACAUGGUUAUUUCCAUGUACGUAUAUAAAUUUGCAAUCGUAUGGCGAUGGACACGCACAAGCCUAGUGA .(((((((...)))))))......((((..(((((((..((((((((((((......))))))..))))))..)))))))((((.....))))......))))........... ( -34.80) >DroYak_CAF1 195790 113 + 1 CUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAUAUAUACAUAUGG-UAUUGCCAUGUGCGUAUAUAAAUAUGCAAUCGUAUGACGAUGGACAUGCAUGAGCCUAGUGA .(((((((...)))))))..(((((.(((((((...((((((..(((((((-.....)))))))..))))))...((((.((((.....)))).).))))))).))))).))). ( -35.60) >consensus CUGCAGUCGUCGAUUGCACCUACAGUGCUAUGCAAGUAAAUAUGCAUAUGGUUAUUGCCAUGUACGUAUAUAAAUUUGCAAUCGUAUGACGAUGGACACGCACAAGCCUAGUGA (((..((((((((((((((.....))))...((((((..((((((((((((......))))))..))))))..))))))))))).)))))..))).(((...........))). (-31.87 = -31.10 + -0.77)

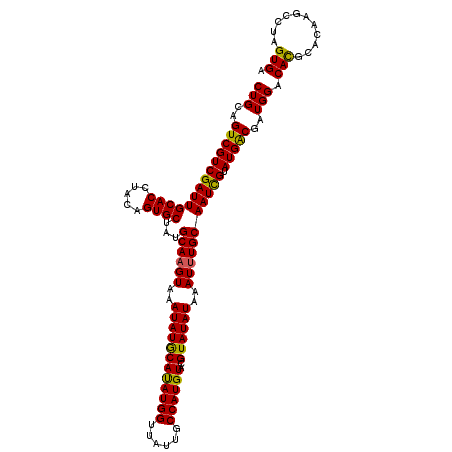
| Location | 12,819,923 – 12,820,037 |
|---|---|
| Length | 114 |
| Sequences | 3 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 92.40 |
| Mean single sequence MFE | -30.13 |
| Consensus MFE | -24.63 |
| Energy contribution | -24.30 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.03 |
| Structure conservation index | 0.82 |
| SVM decision value | 0.00 |
| SVM RNA-class probability | 0.535045 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12819923 114 - 27905053 UCACUAGGUUUGUGCGUGUCCAUCGUCAUACAAUUGCAAAUUUAUAUACGUACAUGGCAAUAACCAUAUGCAUAUUUACUUGCAUAGCACUGUAGGUGCAAUCGACGACUGCAG .(((.......)))..(((...(((((.......(((((.(..((((..(((.((((......)))).)))))))..).)))))..((((.....))))....)))))..))). ( -25.10) >DroSim_CAF1 197536 114 - 1 UCACUAGGCUUGUGCGUGUCCAUCGCCAUACGAUUGCAAAUUUAUAUACGUACAUGGAAAUAACCAUGUGCAUAUUUACUUGCAUAGCACUGUAGGUGCAAUCGACGACUGCAG .......((.((((.(((.....)))))))(((((((............((((((((......))))))))......(((((((......))))))))))))))......)).. ( -34.50) >DroYak_CAF1 195790 113 - 1 UCACUAGGCUCAUGCAUGUCCAUCGUCAUACGAUUGCAUAUUUAUAUACGCACAUGGCAAUA-CCAUAUGUAUAUAUACUUGCAUAGCACUGUAGGUGCAAUCGACGACUGCAG .......((....)).(((...(((((....(((((((....((((((..((.((((.....-)))).))..))))))((((((......))))))))))))))))))..))). ( -30.80) >consensus UCACUAGGCUUGUGCGUGUCCAUCGUCAUACGAUUGCAAAUUUAUAUACGUACAUGGCAAUAACCAUAUGCAUAUUUACUUGCAUAGCACUGUAGGUGCAAUCGACGACUGCAG ............((((.(((..(((.....)((((((............(((.((((......)))).)))......(((((((......))))))))))))))).))))))). (-24.63 = -24.30 + -0.33)

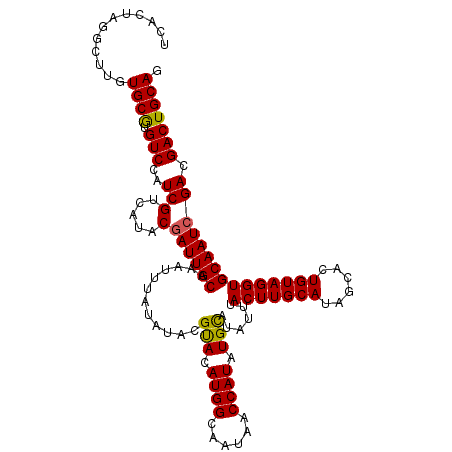
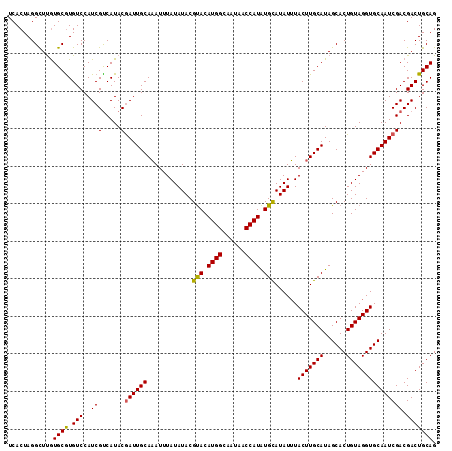
| Location | 12,819,961 – 12,820,077 |
|---|---|
| Length | 116 |
| Sequences | 3 |
| Columns | 116 |
| Reading direction | forward |
| Mean pairwise identity | 91.38 |
| Mean single sequence MFE | -25.05 |
| Consensus MFE | -21.71 |
| Energy contribution | -21.17 |
| Covariance contribution | -0.55 |
| Combinations/Pair | 1.14 |
| Mean z-score | -1.92 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.24 |
| SVM RNA-class probability | 0.648195 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12819961 116 + 27905053 AAUAUGCAUAUGGUUAUUGCCAUGUACGUAUAUAAAUUUGCAAUUGUAUGACGAUGGACACGCACAAACCUAGUGAGAAUUAAUCUAGUGCAUUAAAGCAAUUUUUACAAUAAUAA .(((((.(((((((....))))))).))))).((((.((((...(((((...(((.......(((.......))).......)))..))))).....))))..))))......... ( -22.14) >DroSim_CAF1 197574 116 + 1 AAUAUGCACAUGGUUAUUUCCAUGUACGUAUAUAAAUUUGCAAUCGUAUGGCGAUGGACACGCACAAGCCUAGUGAGAAUUAAUCUAGUGCAUUAAAGCAAUUUUUACAAAAAUAA .(((((.((((((......)))))).))))).((((.((((.((((.....))))......((((.((..((((....))))..)).))))......))))..))))......... ( -26.90) >DroYak_CAF1 195828 115 + 1 UAUAUACAUAUGG-UAUUGCCAUGUGCGUAUAUAAAUAUGCAAUCGUAUGACGAUGGACAUGCAUGAGCCUAGUGAGAAUUAAUCUAGUGCAUUAUAGCAAUUUUUACAAUAAUAA ((((..(((((((-.....)))))))..))))....((((((((((.....)))).....))))))......(((((((((...(((........))).)))))))))........ ( -26.10) >consensus AAUAUGCAUAUGGUUAUUGCCAUGUACGUAUAUAAAUUUGCAAUCGUAUGACGAUGGACACGCACAAGCCUAGUGAGAAUUAAUCUAGUGCAUUAAAGCAAUUUUUACAAUAAUAA .((((((((((((......))))))..)))))).....(((.((((.....))))......((((.((..((((....))))..)).))))......)))................ (-21.71 = -21.17 + -0.55)

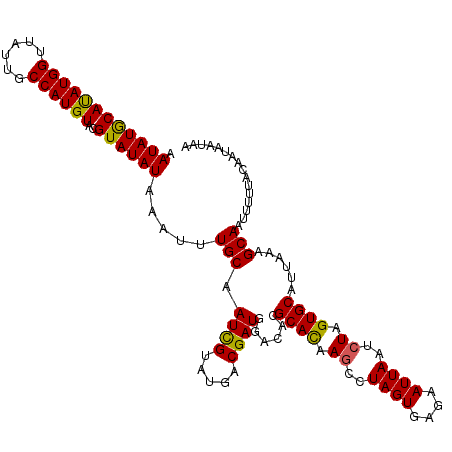
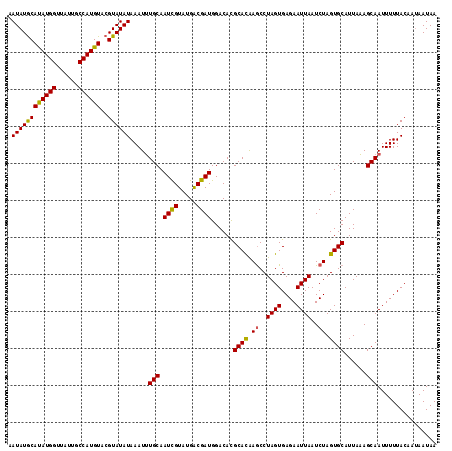
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:59 2006