
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,816,601 – 12,816,801 |
| Length | 200 |
| Max. P | 0.999096 |

| Location | 12,816,601 – 12,816,721 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.75 |
| Mean single sequence MFE | -34.98 |
| Consensus MFE | -30.38 |
| Energy contribution | -30.82 |
| Covariance contribution | 0.44 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.40 |
| Structure conservation index | 0.87 |
| SVM decision value | 0.64 |
| SVM RNA-class probability | 0.808941 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12816601 120 - 27905053 GAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCGCAUAGCCAUCUAAUACACCAGCGUUUGGCAGAAAUACGAUACACAAUGGUGAGCUAUAUAAGCCCACUA ...((((.(((((((((((((.....))))).((((.(((..........))).))))...........)))).)))).))))..............(((((.(((.....))).))))) ( -32.90) >DroSec_CAF1 191457 120 - 1 GAGUUUGGCAAAGCUGCUUUUUGUAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGCGGAAAUACGGUACACAAUGGUGAGCUAUAUAACCCCGCUA .....((((.((((.(((...((((((((((.((..(((.....)))..)).....)).))).)))))..)))))))((.((..(((..((.(((....))).))..)))..)))))))) ( -30.50) >DroSim_CAF1 193371 120 - 1 GAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGCAGAAAUACGCUACACAAUGGUGAGCUAUAUAACCUCGCUA (((...(((((((((.(((((.....))))).)))))))))...........(((((.........(((((..(..((((........))))..)..))))).))))).....))).... ( -33.30) >DroEre_CAF1 200084 120 - 1 GAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUUUAAUACACCAGCGCCUGGCGGAAAUACGCUAUACAAUGGUGAGCUAUAUAAGCAAGCUA ..((.((((((((((.(((((.....))))).)))))))))).))........((((.........(((((..(..(((((......)))))..)..))))).(((.....)))..)))) ( -40.00) >DroYak_CAF1 192660 120 - 1 GAGUUUGGCAAAGCCGCUUUCUGCAUGGAGGAGGUUUUGCUCCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGCGGAAAUACGCUACACAAUGGUGAGCUAUAAAAGCAAGAUA ..((..(((((((((.(((((.....))))).)))))))))..)).............((((....(((((..(..(((((......)))))..)..))))).(((.....))).)))). ( -38.20) >consensus GAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGCGGAAAUACGCUACACAAUGGUGAGCUAUAUAAGCACGCUA ..((.((((((((((.(((((.....))))).)))))))))).)).......(((((.........(((((..(..(((((......)))))..)..))))).)))))............ (-30.38 = -30.82 + 0.44)


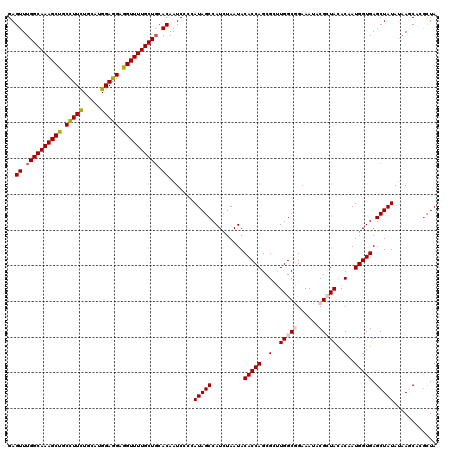
| Location | 12,816,641 – 12,816,761 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 95.00 |
| Mean single sequence MFE | -44.16 |
| Consensus MFE | -42.96 |
| Energy contribution | -43.24 |
| Covariance contribution | 0.28 |
| Combinations/Pair | 1.09 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.62 |
| SVM RNA-class probability | 0.995805 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12816641 120 - 27905053 GGCAUGGCAUGAGCUGUGCAACAUGUAUCUGGCUGAAGGCGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCGCAUAGCCAUCUAAUACACCAGCGUUUGGC .(((((((....)))))))....(((((.((((((...(((..((((((((((((.(((((.....))))).))))))))))...))..))).))))))....)))))((((...)))). ( -43.20) >DroSec_CAF1 191497 120 - 1 GACAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGCGAGUUUGGCAAAGCUGCUUUUUGUAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGC ......((.(((((.((......(((((.((((((.(((.((((.((((((((((.(((((.....))))).)))))))))).))..))))).))))))....)))))...))))))))) ( -39.40) >DroSim_CAF1 193411 120 - 1 GGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGCGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGC .(((((((....)))))))....(((((.((((((.(((.((((.((((((((((.(((((.....))))).)))))))))).))..))))).))))))....)))))((((...)))). ( -47.00) >DroEre_CAF1 200124 120 - 1 GGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGUGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUUUAAUACACCAGCGCCUGGC .(((((((....)))))))....(((((.((((((.(((.((((.((((((((((.(((((.....))))).)))))))))).))..))))).))))))....)))))((((...)))). ( -49.00) >DroYak_CAF1 192700 120 - 1 GGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCCGAGGGUGAGUUUGGCAAAGCCGCUUUCUGCAUGGAGGAGGUUUUGCUCCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGC .(((((((....)))))))....(((((.((((...(((.((((..(((((((((.(((((.....))))).)))))))))..))..)))))...))))....)))))((((...)))). ( -42.20) >consensus GGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGCGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGCUGCACAAUCCCCAUAGCCAUCUAAUACACCAGCGCUUGGC .(((((((....)))))))....(((((.((((((.(((.((((.((((((((((.(((((.....))))).)))))))))).))..))))).))))))....)))))((((...)))). (-42.96 = -43.24 + 0.28)

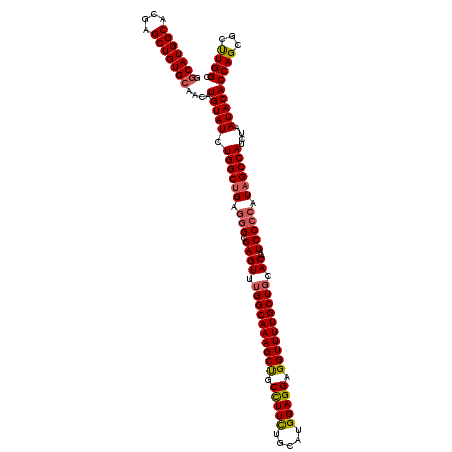
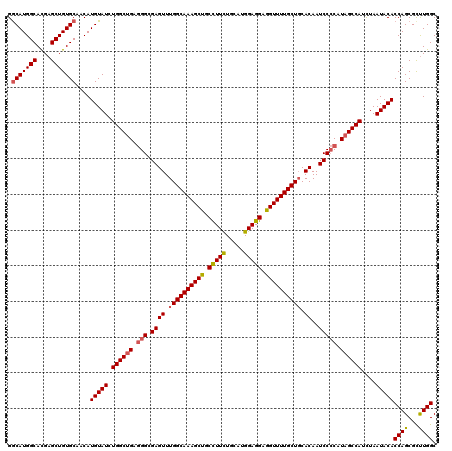
| Location | 12,816,681 – 12,816,801 |
|---|---|
| Length | 120 |
| Sequences | 5 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 92.83 |
| Mean single sequence MFE | -40.52 |
| Consensus MFE | -38.08 |
| Energy contribution | -37.00 |
| Covariance contribution | -1.08 |
| Combinations/Pair | 1.19 |
| Mean z-score | -2.04 |
| Structure conservation index | 0.94 |
| SVM decision value | 3.37 |
| SVM RNA-class probability | 0.999096 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12816681 120 - 27905053 UAACUAUUUAAUAUUCCUUCAGGUUUAUGUCUGACCAGGAGGCAUGGCAUGAGCUGUGCAACAUGUAUCUGGCUGAAGGCGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGC .............((((((((((((((((((((.((....)))).))))))))))((((.....))))...((.((((((.(((((....))))))))))).)).))))))))....... ( -41.80) >DroSec_CAF1 191537 120 - 1 UAAAUAUGUAAUAUUCCUUCAGAUUUAUGUCUGACCAGGAGACAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGCGAGUUUGGCAAAGCUGCUUUUUGUAUGGAGGAGGUUUUGC .......((((..(((((((((((....))))))((((((.(((((((((.....))))..))))).))).((.((((((.(((((....))))))))))).)).))))))))...)))) ( -36.40) >DroSim_CAF1 193451 120 - 1 UAAAUAUGUAAUAUUCCUUCAGGUUUAUGUCUGACCAGGAGGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGCGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGC .......((((..((((((((((((.......)))).(((.(((((((((.....))))..))))).))).((.((((((.(((((....))))))))))).)).))))))))...)))) ( -43.20) >DroEre_CAF1 200164 120 - 1 AAAACAUUUAUAAUUCCUUCAGGUUCAUGUCUGACCAGGAGGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGUGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGC ...............(((((((..(((....)))(((((..(((((((((.....))))..))))).))))))))))))........((((((((.(((((.....))))).)))))))) ( -40.60) >DroYak_CAF1 192740 120 - 1 UAAACAUUUUUAAUUCCUUCAGGUUCAUGUCUGACCAGGAGGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCCGAGGGUGAGUUUGGCAAAGCCGCUUUCUGCAUGGAGGAGGUUUUGC ...................((..(((((.(.((.(((((..(((((((((.....))))..))))).))))).)).).)))))..))((((((((.(((((.....))))).)))))))) ( -40.60) >consensus UAAAUAUUUAAUAUUCCUUCAGGUUUAUGUCUGACCAGGAGGCAUGGCACGAGCUGUGCAACAUGUAUCUGGCUGAGGGCGAGUUUGGCAAAGCUGCCUUCUGCAUGGAGGAGGUUUUGC ...............(((((((..(((....)))(((((..(((((((((.....))))..))))).))))))))))))........((((((((.(((((.....))))).)))))))) (-38.08 = -37.00 + -1.08)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:51 2006