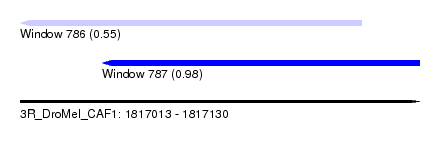
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,817,013 – 1,817,130 |
| Length | 117 |
| Max. P | 0.983162 |

| Location | 1,817,013 – 1,817,113 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 80.53 |
| Mean single sequence MFE | -29.21 |
| Consensus MFE | -17.62 |
| Energy contribution | -18.01 |
| Covariance contribution | 0.39 |
| Combinations/Pair | 1.19 |
| Mean z-score | -1.64 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.04 |
| SVM RNA-class probability | 0.551802 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
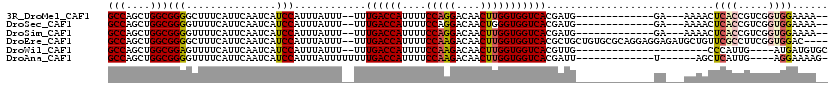
>3R_DroMel_CAF1 1817013 100 - 27905053 GCCAGCUGGCGGGGCUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAGGACAACUUGGUGGUCACGAUG-------------GA---AAAACUCACCGUCGGUGGAAAA-- (((....)))..................(((((......--..((((((....(((((....)))))))))))(((((-------------(.---........)))))))))))...-- ( -29.00) >DroSec_CAF1 127491 100 - 1 GCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAGGACAACUGGGUGGUCACGAUG-------------GA---AAAACUCACCGUCGGUGGAAAA-- .(((.(((((((((((((..........((((((.....--..(((((((...((((.....))))))))))).))))-------------))---))))).).))))))))))....-- ( -33.90) >DroSim_CAF1 130609 100 - 1 GCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAGGACAACUUGGUGGUCACGAUG-------------GA---AAAACUCACCGUCGGUGGAAAA-- .(((.(((((((((((((..........((((((.....--..((((((....(((((....))))))))))).))))-------------))---))))).).))))))))))....-- ( -31.40) >DroEre_CAF1 123653 114 - 1 GCCAGCUGGCGGGGCUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAAGACAACUUGGUGGUCACGCUGCUGUGCGCAGGAGGAGAUGCUGUUCGCCUUCGGUGGAC---- .(((.(.((((((((((((.(((................--..((((((....(((((....)))))))))))..((((.....))))))))))).)))..)))))...).)))..---- ( -36.00) >DroWil_CAF1 129158 92 - 1 GCCAGCUGGCGGAGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUUG----------------------CCCAUUG----AUGAUGUGC (((....))).((((....)))).((((((.........--..((((((....(((((....))))))))))).....----------------------......)----))))).... ( -22.65) >DroAna_CAF1 116605 96 - 1 GCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUUUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGAUU-------------U------AGCUCAUUG----AGGAAAAG- (((....)))....(((((.((((((.................((((((....(((((....))))))))))).((..-------------.------...))))))----))))))).- ( -22.30) >consensus GCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU__UUUGACCAUUUUCCAAGACAACUUGGUGGUCACGAUG_____________GA___AAAACUCACCGUCGGUGGAAAA__ (((....)))(((...............)))............((((((....(((((....)))))))))))............................((((.....))))...... (-17.62 = -18.01 + 0.39)
| Location | 1,817,037 – 1,817,130 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 104 |
| Reading direction | reverse |
| Mean pairwise identity | 86.27 |
| Mean single sequence MFE | -23.62 |
| Consensus MFE | -22.63 |
| Energy contribution | -21.85 |
| Covariance contribution | -0.78 |
| Combinations/Pair | 1.18 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.96 |
| SVM decision value | 1.94 |
| SVM RNA-class probability | 0.983162 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
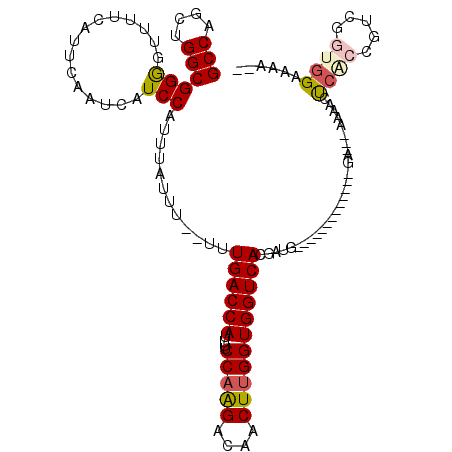
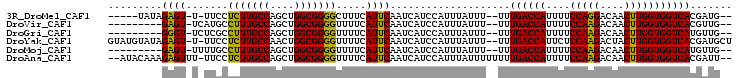
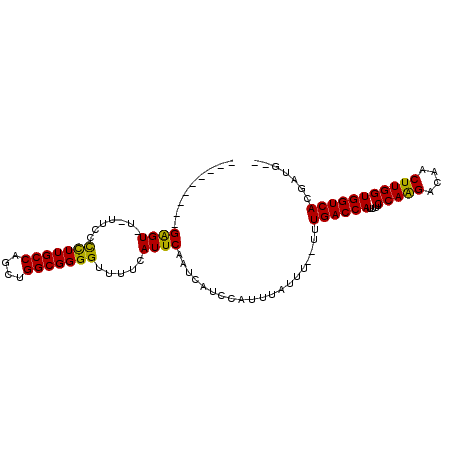
>3R_DroMel_CAF1 1817037 93 - 27905053 -----UAUAGAGU-U-UUCCUCUUGCCAGCUGGCGGGGCUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAGGACAACUUGGUGGUCACGAUG-- -----....((((-.-....(((((((....))))))).....))))................--..((((((....(((((....))))))))))).....-- ( -22.60) >DroVir_CAF1 149115 90 - 1 ---------GAGU-UCAUGCCUUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAAGACAACUUGGUGGUCACGUUG-- ---------((((-....(((((.(((....))))))))....))))................--..((((((....(((((....))))))))))).....-- ( -24.10) >DroGri_CAF1 130521 90 - 1 ---------GGGU-UCUCGCCUUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAAGACAACUUGGUGGUCAUGUUG-- ---------((((-(((((((..........))))))).............))))........--..((((((....(((((....))))))))))).....-- ( -23.61) >DroYak_CAF1 127332 100 - 1 GUAUGUAUAGAGU-U-UUCCUCUUGCCAACUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUCUCCAAGACUACUUGGUGGUCACGAUGCU ....((((.((((-.-....(((((((....))))))).....))))................--..((((((....(((((....)))))))))))..)))). ( -24.70) >DroMoj_CAF1 140650 90 - 1 ---------GAGU-UUUUGCCUUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU--UUUGACCAUUUUCCAAGACAACUUGGUGGUCAUGUUG-- ---------((((-....(((((.(((....))))))))....))))................--..((((((....(((((....))))))))))).....-- ( -24.20) >DroAna_CAF1 116623 99 - 1 --AUACAAAGAGUUU-UUCCUCUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUUUUUUUGACCAUUUUCCAAGACAACUUGGUGGUCACGAUU-- --.......((((..-..((((..(((....))))))).....))))....................((((((....(((((....))))))))))).....-- ( -22.50) >consensus _________GAGU_U_UUCCCCUUGCCAGCUGGCGGGGUUUUCAUUCAAUCAUCCAUUUAUUU__UUUGACCAUUUUCCAAGACAACUUGGUGGUCACGAUG__ .........((((.......(((((((....))))))).....))))....................((((((....(((((....)))))))))))....... (-22.63 = -21.85 + -0.78)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:24 2006