
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,804,616 – 12,804,772 |
| Length | 156 |
| Max. P | 0.681397 |

| Location | 12,804,616 – 12,804,736 |
|---|---|
| Length | 120 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 78.83 |
| Mean single sequence MFE | -42.00 |
| Consensus MFE | -26.54 |
| Energy contribution | -26.85 |
| Covariance contribution | 0.31 |
| Combinations/Pair | 1.48 |
| Mean z-score | -1.53 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.31 |
| SVM RNA-class probability | 0.681397 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
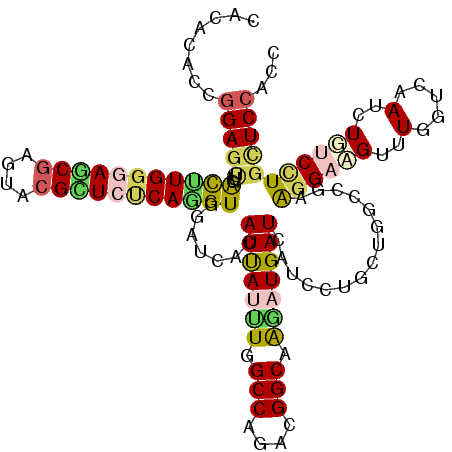
>3R_DroMel_CAF1 12804616 120 - 27905053 CAUACCCCGGAGCUAACGUGGGAGCGUGUCCGUUCUCAAGUGGAUCACAUCAGGUGGCCGGACGGCAGGAUGAUCAUUUUGCUCGCCGAGGGAAGACUGGUGAAUUUGUCCUGUUCCACC ........(((((..((.((((((((....)))))))).))((.((((.....))))))((((((((((((....)))))))(((((.((......)))))))...))))).)))))... ( -42.80) >DroVir_CAF1 219245 120 - 1 CAUACACCGGAGUUAACUUGGGAGCGAGUGCGCUCGCAGGUGGAACAUAUUACUUGGCCAGACGGCAAGAUGAUCAUUCUCCUAGCUGAAGGACGUUUGGUCAAUCUGUCGUGUUCCACC ......((.(((....))).)).(((((....))))).((((((((((((...(((((((((((.......((....))((((......)))))))))))))))...)).)))))))))) ( -48.60) >DroGri_CAF1 304962 120 - 1 CACACACCGGAGAUGACUUGGCAGCAUGUACGCUCUCAAGUCGAUCACAUUAUUUGGCCCGACGGCAAGAUGAUCAUCCUGCUGGCCGAGGGACGUUUGGUUAAUCUGUCCUGCUCCACC ........((((.((((((((.(((......))).))))))))..........((((((....((((.(((....))).))))))))))((((((..(.....)..)))))).))))... ( -40.20) >DroWil_CAF1 248111 120 - 1 CACACUCCGGAGCUUGUCUGGGAACGAGUACGUUCUCAGGUAGAUCACAUUAUUUGGCCAGAUGGCAAAAUGAUUAUCCUGCUUGCCGAGGGUAGACUAGUCAAUCUUUCCUGUUCAACC .......((((((.(..(((((((((....)))))))))..)(((...(((((((.(((....))).))))))).)))..)))..)))((((.(((........))).))))........ ( -33.10) >DroMoj_CAF1 223432 120 - 1 CACACGCCGGAGUUGACCUGGCAGCGAGUGCGCUCCCAGGUGGAUCACAUUAUUUGGCCUGACGGCAAGAUGAUCAUUCUCCUGGCCGAAGGACGUUUGGUAAAUCUGUCCUGCUCCACC .....(((((((((.((((((.((((....)))).)))))).))....(((((((.(((....))).)))))))....)))).)))...((((((..(.....)..))))))........ ( -41.80) >DroAna_CAF1 168190 120 - 1 CAUACGCCGGAAUUGACUUGGGAACGAGUUCGCUCCCAGGUUGACCACAUCAUCUGGCCCGACGGCAGGAUGAUUAUCCUUCUGGCAGAAGGUAGGUUGGUGAAUCUCUCUUGCUCCACA .....((((((((..((((((((.((....)).))))))))..)....(((((((.(((....))).))))))).....)))))))....((((((..((.....))..))))))..... ( -45.50) >consensus CACACACCGGAGUUGACUUGGGAGCGAGUACGCUCUCAGGUGGAUCACAUUAUUUGGCCAGACGGCAAGAUGAUCAUCCUGCUGGCCGAAGGAAGUUUGGUCAAUCUGUCCUGCUCCACC ........(((((..(((((((((((....))))))))))).......(((((((.(((....))).)))))))...............((((((..(.....)..)))))))))))... (-26.54 = -26.85 + 0.31)


| Location | 12,804,656 – 12,804,772 |
|---|---|
| Length | 116 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 83.25 |
| Mean single sequence MFE | -36.52 |
| Consensus MFE | -29.80 |
| Energy contribution | -29.97 |
| Covariance contribution | 0.17 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.82 |
| SVM decision value | -0.07 |
| SVM RNA-class probability | 0.500000 |
| Prediction | OTHER |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12804656 116 - 27905053 ---UAUCAAAAGGUUUUUAACCCACUUUAGAAUGGCUUGCAUACCCCGGAGCUAACGUGGGAGCGUGUCCGUUCUCAAGUGGAUCACAUCAGGUGGCCGGACGGCAGGAUGAUCAUUUU ---........(((.......((((((.((((((((.(((...(((((.......)).))).))).).))))))).))))))....((((.....(((....)))..)))))))..... ( -36.40) >DroSec_CAF1 179506 116 - 1 ---UAUCAAUAGGUUUUUAACACACUUUAGAAUGGCUUGCAUACCCCGGAGCUCACGUGGGAGCGUGUGCGUUCCCAAGUGGACCACAUCAGGUGGCCGGACGGCAGGAUGAUCAUUUU ---.........................((((((((((((.....((((...((((.((((((((....)))))))).)))).((((.....))))))))...)))))....))))))) ( -37.40) >DroSim_CAF1 181438 116 - 1 ---UAUCAAAAGGUUUUUAACCCACUUUAGAAUGGCUUGCAUACCCCGGAGCUAACGUGGGAGCGUGUCCGUUCCCAAGUGGACCACAUCAGGUGGCCGGACGGCAGGAUGAUCAUUUU ---........(((.....)))......((((((((((((.....((((..(((...((((((((....))))))))..))).((((.....))))))))...)))))....))))))) ( -35.20) >DroEre_CAF1 188155 116 - 1 ---UAUUAAACUGAUCUUAAACCCUAUCAGAAUGGCUUGCAUACACCGGAACUAACGUGGGAGCGUGUCCGUUCCCAAGUGGACCACAUCAGGUGGCCGGAUGGCAGGAUGAUCAUUUU ---........(((((......((.........))(((((.....((((..(((...((((((((....))))))))..))).((((.....))))))))...)))))..))))).... ( -36.90) >DroYak_CAF1 178890 116 - 1 ---UACUAAAACGAGUUUCAUCCAUUUCAGAAUGGACUGCAUACCCCGGAGCUAACGUGGGAGCGUGUCCGUUCCCAAGUGGACCACAUCAGGUGGCCGGACGGCAGGAUGAUCAUUUU ---.........((...((.((((((....))))))((((.....((((..(((...((((((((....))))))))..))).((((.....))))))))...))))))...))..... ( -39.40) >DroAna_CAF1 168230 116 - 1 UCCUUUCAAAUCUUAUU---UCCCUUACAGAACAGCCUGCAUACGCCGGAAUUGACUUGGGAACGAGUUCGCUCCCAGGUUGACCACAUCAUCUGGCCCGACGGCAGGAUGAUUAUCCU .................---............((.(((((....((((((...(((((((((.((....)).)))))))))((.....)).))))))......))))).))........ ( -33.80) >consensus ___UAUCAAAAGGUUUUUAACCCACUUCAGAAUGGCUUGCAUACCCCGGAGCUAACGUGGGAGCGUGUCCGUUCCCAAGUGGACCACAUCAGGUGGCCGGACGGCAGGAUGAUCAUUUU ............................((((((((((((.....((((..(((...((((((((....))))))))..))).((((.....))))))))...)))))....))))))) (-29.80 = -29.97 + 0.17)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:45 2006