| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,778,366 – 12,778,473 |
| Length | 107 |
| Max. P | 0.996263 |

| Location | 12,778,366 – 12,778,473 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -34.92 |
| Consensus MFE | -33.78 |
| Energy contribution | -33.20 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.97 |
| SVM decision value | 2.67 |
| SVM RNA-class probability | 0.996263 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12778366 107 + 27905053 CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGAGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCC-GUCAAGUGGGUUGAGUGCCCACACGAUGGCCAUUCCCAUUC-- ........((((((..................((((((((.(((.(((.....))).)))))))))-.))..((((((.....)))))).))))))............-- ( -34.00) >DroPse_CAF1 161892 98 + 1 CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGGGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCC-GCCAAGUGGGUUGAGUGCCCACACGAUGGCCG----------- ........((((((..................((((((((.(((.(((.....))).)))))))))-.))..((((((.....)))))).))))))...----------- ( -36.10) >DroSec_CAF1 153200 101 + 1 CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGAGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCC-GUCAAGUGGGUUGAGUGCCCACACGAUGGUCAUUC-------- ........((((((..................((((((((.(((.(((.....))).)))))))))-.))..((((((.....)))))).))))))......-------- ( -34.20) >DroEre_CAF1 160450 109 + 1 CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGAGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCC-GUCAAGUGGGUAGAGUGCCCACACGAUGGCCAUUCCCAUUCCC ........((((((..................((((((((.(((.(((.....))).)))))))))-.))..((((((.....)))))).)))))).............. ( -33.70) >DroAna_CAF1 140007 103 + 1 CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGAGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCCAGUCAAGUGGGCUGAGUGCCCACACGAUGGCCAUUCC------- ........((((((....................((((((.(((.(((.....))).)))))))))......((((((.....)))))).)))))).......------- ( -35.40) >DroPer_CAF1 169002 98 + 1 CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGGGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCC-GCCAAGUGGGUUGAGUGCCCACACGAUGGCCA----------- ........((((((..................((((((((.(((.(((.....))).)))))))))-.))..((((((.....)))))).))))))...----------- ( -36.10) >consensus CUAUAAAUCCAUCGAAAUAAUGGAUCGGAUAAGAGGCGCGCAUUCGAGAUGAGUUCCAGUCGCGCC_GUCAAGUGGGUUGAGUGCCCACACGAUGGCCAUUC________ ........((((((..................((((((((.(((.(((.....))).)))))))))..))..((((((.....)))))).)))))).............. (-33.78 = -33.20 + -0.58)

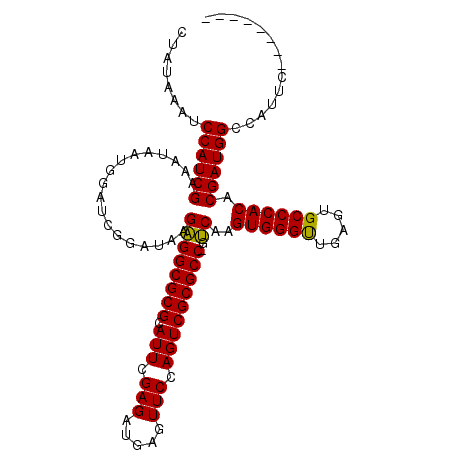

| Location | 12,778,366 – 12,778,473 |
|---|---|
| Length | 107 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 92.17 |
| Mean single sequence MFE | -33.23 |
| Consensus MFE | -28.57 |
| Energy contribution | -28.52 |
| Covariance contribution | -0.06 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.48 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.08 |
| SVM RNA-class probability | 0.572965 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12778366 107 - 27905053 --GAAUGGGAAUGGCCAUCGUGUGGGCACUCAACCCACUUGAC-GGCGCGACUGGAACUCAUCUCGAAUGCGCGCCUCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG --(.((((((.......(((.(((((.......))))).))).-((((((..(.((.......)).)...)))))))))))).)((((((((.....))))))))..... ( -32.30) >DroPse_CAF1 161892 98 - 1 -----------CGGCCAUCGUGUGGGCACUCAACCCACUUGGC-GGCGCGACUGGAACUCAUCUCGAAUGCGCGCCCCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG -----------..((((....(((((.......))))).))))-((((((..(.((.......)).)...))))))........((((((((.....))))))))..... ( -33.30) >DroSec_CAF1 153200 101 - 1 --------GAAUGACCAUCGUGUGGGCACUCAACCCACUUGAC-GGCGCGACUGGAACUCAUCUCGAAUGCGCGCCUCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG --------......((((((.(((((.......)))))..((.-((((((..(.((.......)).)...))))))))..................))))))........ ( -29.20) >DroEre_CAF1 160450 109 - 1 GGGAAUGGGAAUGGCCAUCGUGUGGGCACUCUACCCACUUGAC-GGCGCGACUGGAACUCAUCUCGAAUGCGCGCCUCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG .(((.(((((.......(((.(((((.......))))).))).-((((((..(.((.......)).)...))))))))))))))((((((((.....))))))))..... ( -35.30) >DroAna_CAF1 140007 103 - 1 -------GGAAUGGCCAUCGUGUGGGCACUCAGCCCACUUGACUGGCGCGACUGGAACUCAUCUCGAAUGCGCGCCUCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG -------(((.......(((.((((((.....)))))).)))..((((((..(.((.......)).)...)))))).....)))((((((((.....))))))))..... ( -36.00) >DroPer_CAF1 169002 98 - 1 -----------UGGCCAUCGUGUGGGCACUCAACCCACUUGGC-GGCGCGACUGGAACUCAUCUCGAAUGCGCGCCCCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG -----------..((((....(((((.......))))).))))-((((((..(.((.......)).)...))))))........((((((((.....))))))))..... ( -33.30) >consensus ________GAAUGGCCAUCGUGUGGGCACUCAACCCACUUGAC_GGCGCGACUGGAACUCAUCUCGAAUGCGCGCCUCUUAUCCGAUCCAUUAUUUCGAUGGAUUUAUAG ............((...(((.(((((.......))))).)))..((((((..(.((.......)).)...))))))......))((((((((.....))))))))..... (-28.57 = -28.52 + -0.06)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:40 2006