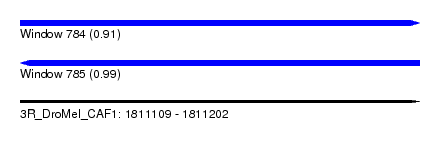
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,811,109 – 1,811,202 |
| Length | 93 |
| Max. P | 0.988635 |

| Location | 1,811,109 – 1,811,202 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | forward |
| Mean pairwise identity | 65.45 |
| Mean single sequence MFE | -32.48 |
| Consensus MFE | -12.32 |
| Energy contribution | -14.52 |
| Covariance contribution | 2.20 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.01 |
| Structure conservation index | 0.38 |
| SVM decision value | 1.04 |
| SVM RNA-class probability | 0.905744 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
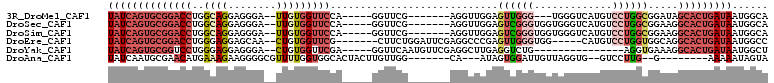
>3R_DroMel_CAF1 1811109 93 + 27905053 UAUCAGUGCGGACCUGGCAGGAGGGA--UUGUGGUUCCA-----GGUUCG-------AGGUUGGAGUUGGG---UGGGUCAUGUCCUGGCGGAUAGCACUGAUAAUGGCA (((((((((....(((.(((((.(((--((.....((((-----(.((((-------....)))).)))))---..)))).).))))).)))...)))))))))...... ( -36.30) >DroSec_CAF1 121730 96 + 1 UAUCAGUGCGGACCUGGCAGGAGGGA--UUGUGGUUCCA-----GGUUCG-------AGGUUGGAGUCGGGUGGUGGGUCAUGUCCUGGCGGAAGGCACUGAUAAUGGCA (((((((((((((((((((.((....--)).))...)))-----))))).-------........((((((..(((...)))..)))))).....)))))))))...... ( -37.00) >DroSim_CAF1 124788 96 + 1 UAUCAGUGCGGACCUGGCAGGAGGGA--UUGUGGUUCCA-----GGUUCG-------AGGUUGGAGUCGGGUGGUGGGUCAUGUCCUGGCGGAAGGCACUGAUAAUGGCA (((((((((((((((((((.((....--)).))...)))-----))))).-------........((((((..(((...)))..)))))).....)))))))))...... ( -37.00) >DroEre_CAF1 117848 96 + 1 UAUCAGUGCGGACCUGGGAGGAGCAA--CUGUGGUUCG-------CUUCUGGAUUCGAGGCCCGAGUUGGGUGG-----CAUGUCCUGGUGGCAGGCACUGAUAAUGGCC ((((((((((((((..(.((......--)).))))))(-------((.((((((..(..((((.....))))..-----)..)))).)).)))..)))))))))...... ( -36.80) >DroYak_CAF1 121162 88 + 1 UAUCAGUGCGGUCCUGGGAGGAGGGA--CUGUGGUUCGA-----GGUUCAAUGUUCGAGGCUUGAGGUCUG---------------AGGUGAAAGGCACUGAUAAUGGCU (((((((((((((((.......))))--))))..(((((-----(.(((.......))).))))))((((.---------------.......)))))))))))...... ( -29.80) >DroAna_CAF1 110576 88 + 1 UAUCAAUGCGAACAUGAAAGAAGGGGCGUUUUGGUGGCACUACUUGUUGG-------CA---AUAGUGGAUUGUUAGGUG--GUCCUUG--G--------AAAAAUAGUA .(((((.(((..(..........)..))).)))))((.((((((((...(-------((---((.....)))))))))))--))))...--.--------.......... ( -18.00) >consensus UAUCAGUGCGGACCUGGCAGGAGGGA__UUGUGGUUCCA_____GGUUCG_______AGGUUGGAGUUGGGUGGUGGGUCAUGUCCUGGCGGAAGGCACUGAUAAUGGCA ((((((((((((((..((((........)))))))))............................((((((.............)))))).....)))))))))...... (-12.32 = -14.52 + 2.20)
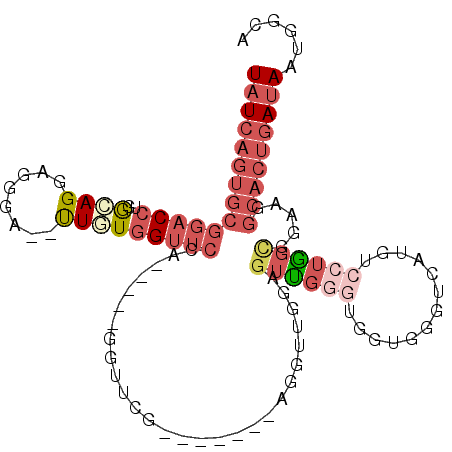
| Location | 1,811,109 – 1,811,202 |
|---|---|
| Length | 93 |
| Sequences | 6 |
| Columns | 110 |
| Reading direction | reverse |
| Mean pairwise identity | 65.45 |
| Mean single sequence MFE | -21.14 |
| Consensus MFE | -9.37 |
| Energy contribution | -10.98 |
| Covariance contribution | 1.61 |
| Combinations/Pair | 1.25 |
| Mean z-score | -2.39 |
| Structure conservation index | 0.44 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988635 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1811109 93 - 27905053 UGCCAUUAUCAGUGCUAUCCGCCAGGACAUGACCCA---CCCAACUCCAACCU-------CGAACC-----UGGAACCACAA--UCCCUCCUGCCAGGUCCGCACUGAUA ......(((((((((..(((....))).........---..............-------.(.(((-----(((........--.........)))))).)))))))))) ( -24.03) >DroSec_CAF1 121730 96 - 1 UGCCAUUAUCAGUGCCUUCCGCCAGGACAUGACCCACCACCCGACUCCAACCU-------CGAACC-----UGGAACCACAA--UCCCUCCUGCCAGGUCCGCACUGAUA ......(((((((((...((..(((((..............(((........)-------))....-----.(((.......--))).)))))...))...))))))))) ( -24.30) >DroSim_CAF1 124788 96 - 1 UGCCAUUAUCAGUGCCUUCCGCCAGGACAUGACCCACCACCCGACUCCAACCU-------CGAACC-----UGGAACCACAA--UCCCUCCUGCCAGGUCCGCACUGAUA ......(((((((((...((..(((((..............(((........)-------))....-----.(((.......--))).)))))...))...))))))))) ( -24.30) >DroEre_CAF1 117848 96 - 1 GGCCAUUAUCAGUGCCUGCCACCAGGACAUG-----CCACCCAACUCGGGCCUCGAAUCCAGAAG-------CGAACCACAG--UUGCUCCUCCCAGGUCCGCACUGAUA ......(((((((((..(((....(((..((-----...(((.....)))...))..)))((.((-------(((.......--))))).))....)))..))))))))) ( -28.10) >DroYak_CAF1 121162 88 - 1 AGCCAUUAUCAGUGCCUUUCACCU---------------CAGACCUCAAGCCUCGAACAUUGAACC-----UCGAACCACAG--UCCCUCCUCCCAGGACCGCACUGAUA ......(((((((((.........---------------.............((((..........-----))))......(--(((.........)))).))))))))) ( -18.00) >DroAna_CAF1 110576 88 - 1 UACUAUUUUU--------C--CAAGGAC--CACCUAACAAUCCACUAU---UG-------CCAACAAGUAGUGCCACCAAAACGCCCCUUCUUUCAUGUUCGCAUUGAUA ..........--------.--.((((..--............((((((---(.-------......))))))).............))))...(((((....)).))).. ( -8.13) >consensus UGCCAUUAUCAGUGCCUUCCGCCAGGACAUGACCCACCACCCAACUCCAACCU_______CGAACC_____UGGAACCACAA__UCCCUCCUGCCAGGUCCGCACUGAUA ......(((((((((...((..(((((.............................................................)))))...))...))))))))) ( -9.37 = -10.98 + 1.61)

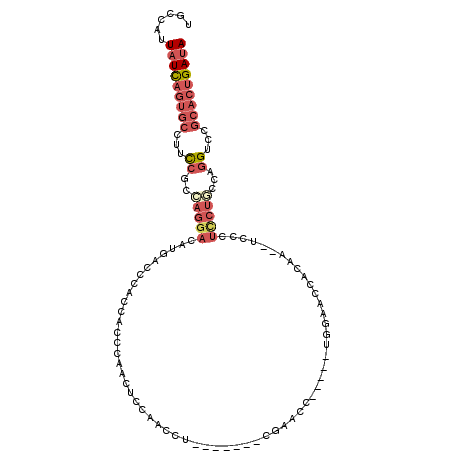
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:22 2006