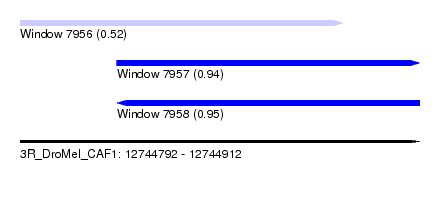
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,744,792 – 12,744,912 |
| Length | 120 |
| Max. P | 0.947738 |

| Location | 12,744,792 – 12,744,889 |
|---|---|
| Length | 97 |
| Sequences | 6 |
| Columns | 103 |
| Reading direction | forward |
| Mean pairwise identity | 78.32 |
| Mean single sequence MFE | -24.28 |
| Consensus MFE | -14.44 |
| Energy contribution | -14.25 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.12 |
| Mean z-score | -1.46 |
| Structure conservation index | 0.59 |
| SVM decision value | -0.03 |
| SVM RNA-class probability | 0.517422 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12744792 97 + 27905053 GCCAACCA------GCACAAACACAUUCGAAUAAGACUCACUCACACAGUGGCAAGCUGUGCAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUC ((..((((------(((((((.....((......))....((..((((((.....))))))..))...)))))))))))..))....(((........))).. ( -27.20) >DroPse_CAF1 124204 89 + 1 GGCAACCA------ACACAAGCACAUAUAAAUUACACUCACUCAUACAGCUGCCAAUUGGAGUUUGUGGGUGUGUUGGUGAGCA----CAGAGUC----UCUC .((.((((------((((.......((.....)).....(((((((.((((.((....)))))))))))))))))))))..)).----.......----.... ( -23.70) >DroSim_CAF1 121593 103 + 1 GCCAACCAGCACCAGCACAAACACAUUCAAAUAAGACUCACUCACACAGCGGCAAGUGGUGAAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUC ((.......((((((((((((.....((......)).((((.(((..........)))))))......)))))))))))).....))(((........))).. ( -24.10) >DroEre_CAF1 126671 97 + 1 GCCAACCA------GCACAAACACAUUCAAAUCAGACUCACUCACACAGCGGCAAGCUGUAAAAGGCAUUUGUGUUGGUGAGCAGGCGAAGAGUUUCAUGCUC (((.((((------(((((((.............((.....))..(((((.....)))))........))))))))))).....)))...((((.....)))) ( -27.60) >DroYak_CAF1 116625 97 + 1 GCCAACCA------GCACAAACACAUUCAAAUCAGACUCACUCACACAGCGGCAAGCUGUAAAAGGCACUUGUGUUGGUGAGCAAGCAAAGAGAUCCAAUCUC ((..((((------((((((..............((.....))..(((((.....))))).........))))))))))..)).......(((((...))))) ( -25.80) >DroAna_CAF1 106604 96 + 1 GCCAGCCA------GCACAAACACUUUCAAAUCAGACUCACCAAUACAACCGCAAACUCUA-GAUGCAAGUGUGUUAGUGAGCAAGCGAAGAGUAUGAUUCUC ((......------)).............((((((.(((((.((((((...(((.......-..)))...)))))).))))))..((.....)).)))))... ( -17.30) >consensus GCCAACCA______GCACAAACACAUUCAAAUCAGACUCACUCACACAGCGGCAAGCUGUAAAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUC ....................................((((((.(((((...((............))...))))).))))))........(((.......))) (-14.44 = -14.25 + -0.19)

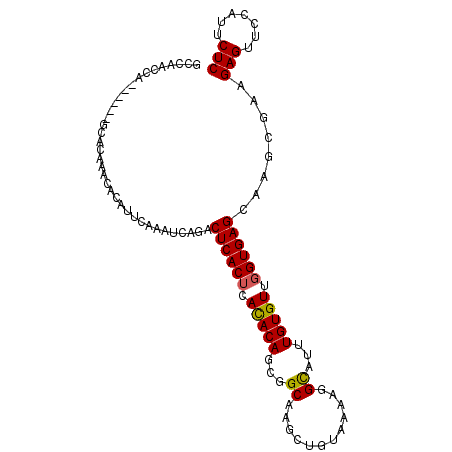

| Location | 12,744,821 – 12,744,912 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -28.37 |
| Consensus MFE | -19.34 |
| Energy contribution | -19.82 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.23 |
| Mean z-score | -2.28 |
| Structure conservation index | 0.68 |
| SVM decision value | 1.35 |
| SVM RNA-class probability | 0.944743 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12744821 91 + 27905053 ACUCACUCACACAGUGGCAAGCUGUGCAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUCUGCUUCG-AAGUACGAAGAGAGAA--------- .((((((.((((((((.(..((...))..).)).)))))).))))))................((((((.(((((-.....)))))))))))--------- ( -29.10) >DroSec_CAF1 120029 91 + 1 ACUCACUCACACAGCGGCAAGCGGUGAAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUCU-CUUCGUAAGUAAGAAGAGGGAA--------- .((((((.(((((((.....))(.(....).)...))))).))))))................((((((-((((........))))))))))--------- ( -29.90) >DroSim_CAF1 121628 92 + 1 ACUCACUCACACAGCGGCAAGUGGUGAAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUCUGCUUCGUAAGUAGGAAGAGAGAA--------- ((((.(((((.(((((.((((((.(....).))))))))))))))))(....)..))))....((((((.((((........))))))))))--------- ( -30.30) >DroEre_CAF1 126700 91 + 1 ACUCACUCACACAGCGGCAAGCUGUAAAAGGCAUUUGUGUUGGUGAGCAGGCGAAGAGUUUCAUGCUCUGCCUU-UCAGUGAGAAGAGAGAA--------- .(((.(((((.(((((.((((.(((.....)))))))))))).((((.((((..(((((.....))))))))))-))))))))..)))....--------- ( -30.70) >DroYak_CAF1 116654 91 + 1 ACUCACUCACACAGCGGCAAGCUGUAAAAGGCACUUGUGUUGGUGAGCAAGCAAAGAGAUCCAAUCUCUGUUUU-UAGGUGAGAAGAGAGAA--------- .(((.((((((((((.....))))).((((((.(((((........)))))...((((((...)))))))))))-)..)))))..)))....--------- ( -27.70) >DroAna_CAF1 106633 100 + 1 ACUCACCAAUACAACCGCAAACUCUA-GAUGCAAGUGUGUUAGUGAGCAAGCGAAGAGUAUGAUUCUCCGAAUAUUCAGAUAGGAAAGAGAAUUCGAGAAG .(((((.((((((...(((.......-..)))...)))))).)))))((.((.....)).)).(((((.((((.(((..........))).))))))))). ( -22.50) >consensus ACUCACUCACACAGCGGCAAGCUGUAAAAGGCAUUUGUGUUGGUGAGCAAGCGAAGAGUUCCAUUCUCUGCUUCGUAAGUAAGAAGAGAGAA_________ .((((((.(((((...((............))...))))).)))))).................(((((.((((........))))))))).......... (-19.34 = -19.82 + 0.47)

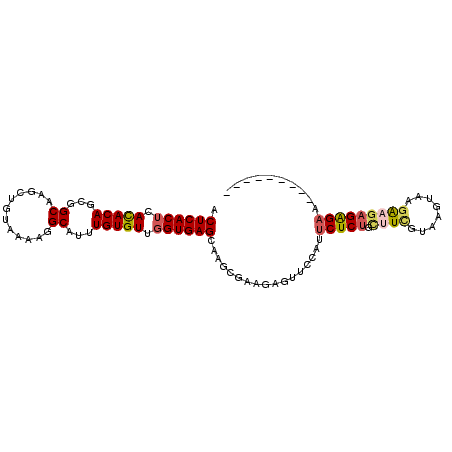
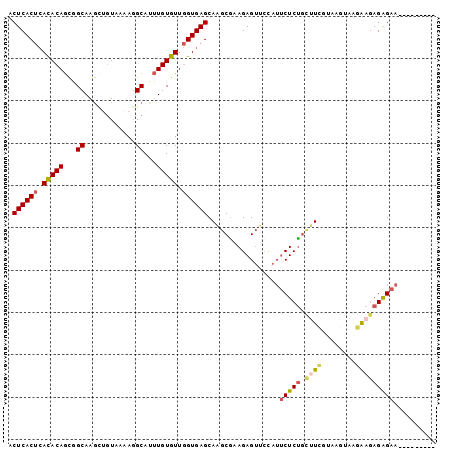
| Location | 12,744,821 – 12,744,912 |
|---|---|
| Length | 91 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | reverse |
| Mean pairwise identity | 79.21 |
| Mean single sequence MFE | -25.50 |
| Consensus MFE | -15.66 |
| Energy contribution | -16.13 |
| Covariance contribution | 0.47 |
| Combinations/Pair | 1.10 |
| Mean z-score | -2.49 |
| Structure conservation index | 0.61 |
| SVM decision value | 1.37 |
| SVM RNA-class probability | 0.947738 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12744821 91 - 27905053 ---------UUCUCUCUUCGUACUU-CGAAGCAGAGAAUGGAACUCUUCGCUUGCUCACCAACACAAAUGCCUUGCACAGCUUGCCACUGUGUGAGUGAGU ---------(((((((((((.....-))))).))))))...............((((((..(((((...((...((...))..))...)))))..)))))) ( -27.80) >DroSec_CAF1 120029 91 - 1 ---------UUCCCUCUUCUUACUUACGAAG-AGAGAAUGGAACUCUUCGCUUGCUCACCAACACAAAUGCCUUUCACCGCUUGCCGCUGUGUGAGUGAGU ---------(((.((((((........))))-)).)))...........(((..(((((((.(.(((..((........)))))..).)).)))))..))) ( -23.10) >DroSim_CAF1 121628 92 - 1 ---------UUCUCUCUUCCUACUUACGAAGCAGAGAAUGGAACUCUUCGCUUGCUCACCAACACAAAUGCCUUUCACCACUUGCCGCUGUGUGAGUGAGU ---------((((((((((........)))).))))))...........(((..(((((((.(.(((.((........)).)))..).)).)))))..))) ( -23.70) >DroEre_CAF1 126700 91 - 1 ---------UUCUCUCUUCUCACUGA-AAGGCAGAGCAUGAAACUCUUCGCCUGCUCACCAACACAAAUGCCUUUUACAGCUUGCCGCUGUGUGAGUGAGU ---------....(((..(((((.((-(((((((((((((((....))))..)))))...........))))))))(((((.....)))))))))).))). ( -30.10) >DroYak_CAF1 116654 91 - 1 ---------UUCUCUCUUCUCACCUA-AAAACAGAGAUUGGAUCUCUUUGCUUGCUCACCAACACAAGUGCCUUUUACAGCUUGCCGCUGUGUGAGUGAGU ---------...(((..((((.....-......))))..))).......(((..(((((...((....))......(((((.....))))))))))..))) ( -23.20) >DroAna_CAF1 106633 100 - 1 CUUCUCGAAUUCUCUUUCCUAUCUGAAUAUUCGGAGAAUCAUACUCUUCGCUUGCUCACUAACACACUUGCAUC-UAGAGUUUGCGGUUGUAUUGGUGAGU .(((((((((..((..........))..)))).)))))...............(((((((((.(((.(((((..-.......))))).))).))))))))) ( -25.10) >consensus _________UUCUCUCUUCUUACUUACGAAGCAGAGAAUGGAACUCUUCGCUUGCUCACCAACACAAAUGCCUUUCACAGCUUGCCGCUGUGUGAGUGAGU ............................((((((((.......))))..))))((((((..(((((...((............))...)))))..)))))) (-15.66 = -16.13 + 0.47)


Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:33 2006