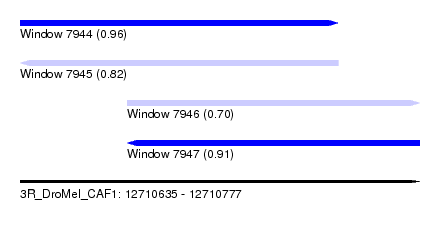
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,710,635 – 12,710,777 |
| Length | 142 |
| Max. P | 0.959515 |

| Location | 12,710,635 – 12,710,748 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | forward |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -45.18 |
| Consensus MFE | -40.72 |
| Energy contribution | -41.13 |
| Covariance contribution | 0.42 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.74 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.50 |
| SVM RNA-class probability | 0.959515 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12710635 113 + 27905053 GGCUUUGGACAGGCUGGAUGUGCGCAGCUUGGCAUAUUGAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAUUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAA ((((((((.(((((((........))))))).(((....)))..))(((((......)))...(((((((.(((((....))))).)))))))..)).))))))......... ( -45.50) >DroSec_CAF1 86373 113 + 1 GGCUUUGGACAGGCUGGAUGUGCGCGGCUUGGCAUAUUGAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAUUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAA (((((((((...((..(...(((((((.....(((....)))..)))).)))..)..))))).(((((((.(((((....))))).))))))).....))))))......... ( -46.70) >DroSim_CAF1 87538 113 + 1 GGCUUUGGACAGGCUGGAUGUGCGCGGCUUGGCAUAUUGAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAUUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAA (((((((((...((..(...(((((((.....(((....)))..)))).)))..)..))))).(((((((.(((((....))))).))))))).....))))))......... ( -46.70) >DroEre_CAF1 91761 113 + 1 GGCUUUGGACAGGCUGGAUGUGCGCGGCUUGGCAUAUUGAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAAUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUGUCGA .......(((((((.((.(.(((((((((((((((....)))..))).)))....(((((((.(((((((...))).))))..))))))).))).))).)..))))))))).. ( -47.30) >DroYak_CAF1 81228 113 + 1 GGCUUUGGACAGGCUGGAUGUGCGCGGCUUGGCAUAUUGAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAAUGCCAACUGGCGAAGCCACCGCAGCAGAAGCCGCUUGGCAA (((((((((...((..(...(((((((.....(((....)))..)))).)))..)..))))).(((((((..((((....))))..))))))).....))))))......... ( -45.30) >DroAna_CAF1 72950 113 + 1 GGCUGUGACCAGGCUGGAUGUGUGUGGCGUGGCAUAUUGAUGAACCGCAACAAAUUGGCUACACGGUGGCAAUGCCAACUGCCGGAGCCACAGAAGCAGAAGCCGUAUGUCAA .....((((..((((...(((.(((((((((((...(((.((.....)).)))....))))).(((((((...)))....))))..))))))...)))..))))....)))). ( -39.60) >consensus GGCUUUGGACAGGCUGGAUGUGCGCGGCUUGGCAUAUUGAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAAUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAA ((((((((...(((..(...(((((((.....(((....)))..)))).)))..)..))))).(((((((..((((....))))..))))))).....))))))......... (-40.72 = -41.13 + 0.42)

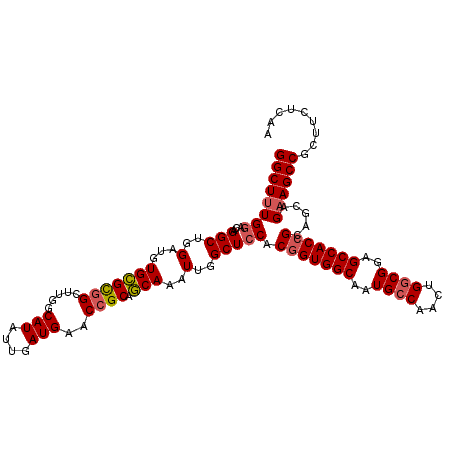
| Location | 12,710,635 – 12,710,748 |
|---|---|
| Length | 113 |
| Sequences | 6 |
| Columns | 113 |
| Reading direction | reverse |
| Mean pairwise identity | 93.92 |
| Mean single sequence MFE | -42.49 |
| Consensus MFE | -36.20 |
| Energy contribution | -36.95 |
| Covariance contribution | 0.75 |
| Combinations/Pair | 1.06 |
| Mean z-score | -1.88 |
| Structure conservation index | 0.85 |
| SVM decision value | 0.67 |
| SVM RNA-class probability | 0.818084 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12710635 113 - 27905053 UUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUCAAUAUGCCAAGCUGCGCACAUCCAGCCUGUCCAAAGCC (((.((.((((((((....((((((((...(((....)))...))))))))))))))....(((.(((((((((....)))...)))))))))......))...))))).... ( -42.10) >DroSec_CAF1 86373 113 - 1 UUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUCAAUAUGCCAAGCCGCGCACAUCCAGCCUGUCCAAAGCC (((.((.((((((((....((((((((...(((....)))...))))))))))))))....(((.(((((((((....)))...)))))))))......))...))))).... ( -44.30) >DroSim_CAF1 87538 113 - 1 UUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUCAAUAUGCCAAGCCGCGCACAUCCAGCCUGUCCAAAGCC (((.((.((((((((....((((((((...(((....)))...))))))))))))))....(((.(((((((((....)))...)))))))))......))...))))).... ( -44.30) >DroEre_CAF1 91761 113 - 1 UCGACAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAUUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUCAAUAUGCCAAGCCGCGCACAUCCAGCCUGUCCAAAGCC ..((((.((((((((....((((((((...(((....)))...))))))))))))))....(((.(((((((((....)))...)))))))))......)).))))....... ( -48.10) >DroYak_CAF1 81228 113 - 1 UUGCCAAGCGGCUUCUGCUGCGGUGGCUUCGCCAGUUGGCAUUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUCAAUAUGCCAAGCCGCGCACAUCCAGCCUGUCCAAAGCC (((.((.((((((((....((((((((...(((....)))...))))))))))))))....(((.(((((((((....)))...)))))))))......)).))..))).... ( -43.80) >DroAna_CAF1 72950 113 - 1 UUGACAUACGGCUUCUGCUUCUGUGGCUCCGGCAGUUGGCAUUGCCACCGUGUAGCCAAUUUGUUGCGGUUCAUCAAUAUGCCACGCCACACACAUCCAGCCUGGUCACAGCC .((((....((((........((((((...(((((.((((...)))).).((((((......))))))...........))))..)))))).......))))..))))..... ( -32.36) >consensus UUGACAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUCAAUAUGCCAAGCCGCGCACAUCCAGCCUGUCCAAAGCC .......((((((((....((((((((...(((....)))...))))))))))))))....(((.(((((((((....)))...)))))))))......))............ (-36.20 = -36.95 + 0.75)

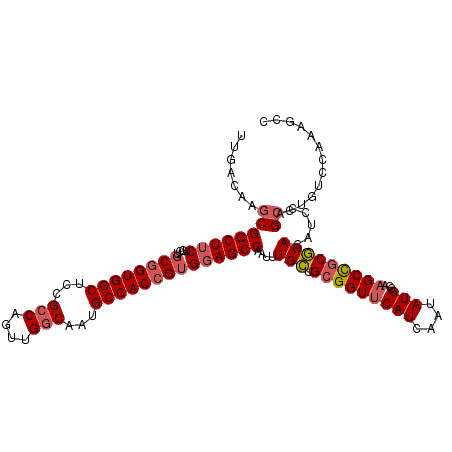
| Location | 12,710,673 – 12,710,777 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 88.21 |
| Mean single sequence MFE | -36.03 |
| Consensus MFE | -31.74 |
| Energy contribution | -31.97 |
| Covariance contribution | 0.22 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.54 |
| Structure conservation index | 0.88 |
| SVM decision value | 0.35 |
| SVM RNA-class probability | 0.701464 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
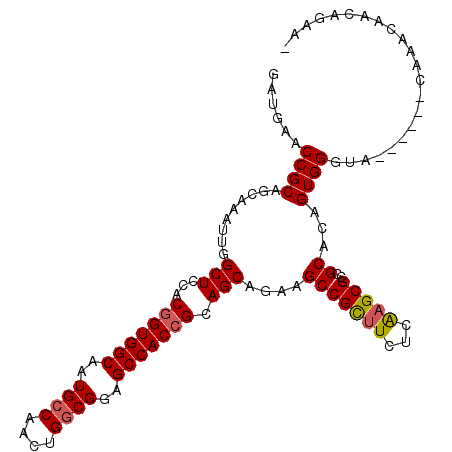
>3R_DroMel_CAF1 12710673 104 + 27905053 GAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAUUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAAGCGCCGCACAGUGGAUA------UAAAUAACAGAA- ......((((.........(((...(((((((.(((((....))))).))))))).)))....((((((.....))))..))...))))...------............- ( -35.40) >DroSec_CAF1 86411 104 + 1 GAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAUUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAAGCGCCGCACAGUGGGUA------CGAACAACAGAA- ......((((.........(((...(((((((.(((((....))))).))))))).)))....((((((.....))))..))...))))...------............- ( -35.60) >DroSim_CAF1 87576 104 + 1 GAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAUUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAAGCGCCGCACAGUGGGUA------CAAUCAACAGAA- (((...((((.........(((...(((((((.(((((....))))).))))))).)))....((((((.....))))..))...))))...------..))).......- ( -37.20) >DroEre_CAF1 91799 104 + 1 GAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAAUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUGUCGAGCGCCGCACAGUGGGCA------CAAGCAACAUAC- .(((..............(((((..(((((((..((((....))))..))))))).....).))))((((((...((.((((...)))))))------)))))..)))..- ( -39.80) >DroYak_CAF1 81266 104 + 1 GAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAAUGCCAACUGGCGAAGCCACCGCAGCAGAAGCCGCUUGGCAAGCGCCGCACAGUGGGUU------GAAACAACAGGA- ......((........(..((.((((((((((..((((....))))..))))))((.((....(((....)))....)).))...)))))).------.).......)).- ( -38.56) >DroAna_CAF1 72988 110 + 1 GAUGAACCGCAACAAAUUGGCUACACGGUGGCAAUGCCAACUGCCGGAGCCACAGAAGCAGAAGCCGUAUGUCAAGCGCCGCACUGUGGUUUUUGGUUUAAAUA-UAGAAC ..((((((((..(....(((((...(((((((...)))....)))).)))))..)..))(((((((((((((........))).))))))))))))))))....-...... ( -29.60) >consensus GAUGAACCGCAGCAAAUUGGCUCCACGGUGGCAAUGCCAACUGGCGGAGCCACCGCAGCAGAAGCCGCUUCUCAAGCGCCGCACAGUGGGUA______CAAACAACAGAA_ ......((((.........(((...(((((((..((((....))))..))))))).)))....(((((((...)))))..))...))))...................... (-31.74 = -31.97 + 0.22)


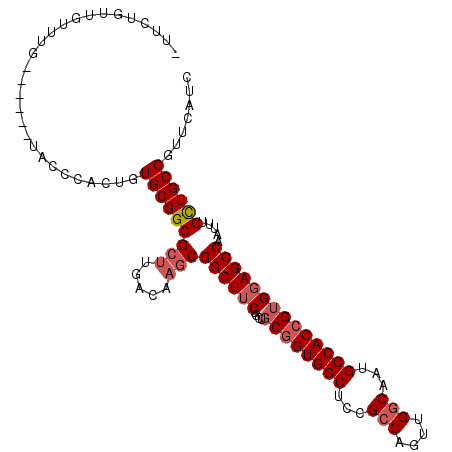
| Location | 12,710,673 – 12,710,777 |
|---|---|
| Length | 104 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | reverse |
| Mean pairwise identity | 88.21 |
| Mean single sequence MFE | -40.58 |
| Consensus MFE | -36.59 |
| Energy contribution | -37.45 |
| Covariance contribution | 0.86 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.79 |
| Structure conservation index | 0.90 |
| SVM decision value | 1.06 |
| SVM RNA-class probability | 0.908925 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12710673 104 - 27905053 -UUCUGUUAUUUA------UAUCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUC -............------.........(((((((((.....)))((((((....((((((((...(((....)))...)))))))))))))).....))))))....... ( -41.80) >DroSec_CAF1 86411 104 - 1 -UUCUGUUGUUCG------UACCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUC -...........(------.(((......((((((((.....)))((((((....((((((((...(((....)))...)))))))))))))).....)))))))).)... ( -41.90) >DroSim_CAF1 87576 104 - 1 -UUCUGUUGAUUG------UACCCACUGUGCGGCGCUUGAGAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUC -..........((------.(((......((((((((.....)))((((((....((((((((...(((....)))...)))))))))))))).....)))))))).)).. ( -42.30) >DroEre_CAF1 91799 104 - 1 -GUAUGUUGCUUG------UGCCCACUGUGCGGCGCUCGACAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAUUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUC -((.(((((...(------(((((.....).))))).))))).))((((((....((((((((...(((....)))...)))))))))))))).................. ( -44.10) >DroYak_CAF1 81266 104 - 1 -UCCUGUUGUUUC------AACCCACUGUGCGGCGCUUGCCAAGCGGCUUCUGCUGCGGUGGCUUCGCCAGUUGGCAUUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUC -.((.((((...)------))).......((((((((.....)))((((((....((((((((...(((....)))...)))))))))))))).....)))))))...... ( -42.80) >DroAna_CAF1 72988 110 - 1 GUUCUA-UAUUUAAACCAAAAACCACAGUGCGGCGCUUGACAUACGGCUUCUGCUUCUGUGGCUCCGGCAGUUGGCAUUGCCACCGUGUAGCCAAUUUGUUGCGGUUCAUC ......-......((((.....((((((.((((.(((........)))..))))..))))))...(((((((((((..(((......)))))))).)))))).)))).... ( -30.60) >consensus _UUCUGUUGUUUG______UACCCACUGUGCGGCGCUUGACAAGCGGCUUCUGCUGCGGUGGCUCCGCCAGUUGGCAAUGCCACCGUGGAGCCAAUUUGCUGCGGUUCAUC ............................(((((((((.....)))((((((....((((((((...(((....)))...)))))))))))))).....))))))....... (-36.59 = -37.45 + 0.86)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:23 2006