
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,697,237 – 12,697,377 |
| Length | 140 |
| Max. P | 0.832200 |

| Location | 12,697,237 – 12,697,327 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 101 |
| Reading direction | forward |
| Mean pairwise identity | 80.34 |
| Mean single sequence MFE | -21.23 |
| Consensus MFE | -15.19 |
| Energy contribution | -15.22 |
| Covariance contribution | 0.03 |
| Combinations/Pair | 1.24 |
| Mean z-score | -1.44 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.38 |
| SVM RNA-class probability | 0.711034 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12697237 90 + 27905053 UGGCCCUGCCUACUGCCGGCUU-----------UUGUUGGCGGCAGUAAAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUG .(((...)))((((((((.(..-----------.....).))))))))............((((....((((((((......))))))))))))....... ( -27.30) >DroVir_CAF1 76753 93 + 1 GC--------AGCGGCUGGCUUUGUUAUUCUUGUUACUGGCGGCAGUAAAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUG ((--------((((..((((....(((((.((((.....)))).))))).....))))..))))...(((((((((......)))))))))))........ ( -21.70) >DroPse_CAF1 74631 81 + 1 A---------CGCUGACGGCUC-----------UUGUUGGCGGUAGUAAAAUUCGUUAAAUGCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUG .---------(((..(((....-----------.)))..)))..................(((....(((((((((......))))))))))))....... ( -17.40) >DroGri_CAF1 125191 93 + 1 GC--------GGCGGCUGACUUUGUUAUUCUUGUUACUGGCGGCAGUAAAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUG ((--------((((..((((....(((((.((((.....)))).))))).....))))..))))...(((((((((......)))))))))))........ ( -22.20) >DroAna_CAF1 60363 82 + 1 CG--------UUCUGCCGGCUU-----------UUGUUGGCGGUAGUAAAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUG ..--------..((((((((..-----------..)))))))).................((((....((((((((......))))))))))))....... ( -21.40) >DroPer_CAF1 81060 81 + 1 A---------CGCUGACGGCUC-----------UUGUUGGCGGUAGUAAAAUUCGUUAAAUGCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUG .---------(((..(((....-----------.)))..)))..................(((....(((((((((......))))))))))))....... ( -17.40) >consensus A_________CGCUGCCGGCUU___________UUGUUGGCGGCAGUAAAAUUCGUUAAAUGCUCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUG ...........(((((((.(..................).)))))))....................(((((((((......))))))))).......... (-15.19 = -15.22 + 0.03)


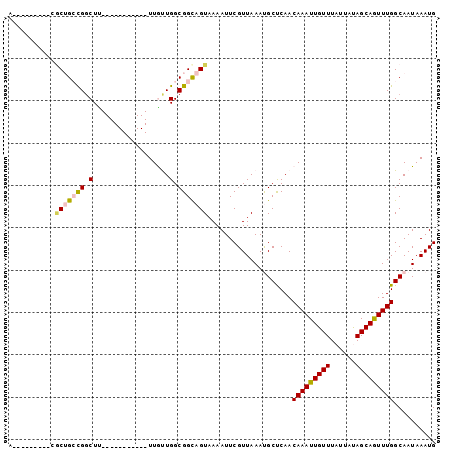
| Location | 12,697,258 – 12,697,367 |
|---|---|
| Length | 109 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 85.40 |
| Mean single sequence MFE | -28.70 |
| Consensus MFE | -24.58 |
| Energy contribution | -24.25 |
| Covariance contribution | -0.33 |
| Combinations/Pair | 1.21 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.86 |
| SVM decision value | 0.72 |
| SVM RNA-class probability | 0.832200 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12697258 109 + 27905053 U-----------UUGUUGGCGGCAGUAAAAUUCGUUAAAUGCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGUUGCCACCGCACUUUUC .-----------.(((((((((((.((((((..(((((..(((...(((((((((......)))))))))(((.....))))))..)))))..))))).).))))))))..)))...... ( -32.10) >DroVir_CAF1 76766 120 + 1 UUGUUAUUCUUGUUACUGGCGGCAGUAAAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUGCGGCUAUUAGCUCAUUUUAGGUGUUGCCACCGCACUUUUC ..........(((...((((((((.((((((..(((((..(((...(((((((((......)))))))))(((.....))))))..)))))..))))).).))))))))..)))...... ( -29.30) >DroGri_CAF1 125204 120 + 1 UUGUUAUUCUUGUUACUGGCGGCAGUAAAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUGCGGCCAUUAGCUCAUUGUAGGUGUUGCCACCGCACACUUA ...........((((.((((.(((........((.....)).....(((((((((......)))))))))........))).)))).))))......(((((((.((....))))))))) ( -30.30) >DroWil_CAF1 99129 109 + 1 G-----------CUACUGACGGCAGUAAAAUUCGUUAAAUGCUUAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCCAUUAACUCAUUUUAGGUGUUGCCACCGCACUUUUC (-----------((((((....)))))......(((((..(((...(((((((((......)))))))))(((.....))))))..)))))........((((....))))))....... ( -25.40) >DroMoj_CAF1 86020 120 + 1 UUGUUAUUCUUGUUACUGGUGGCAGUAAAAUUCGUUAAACGCUAAACAAAUUGUUUAUUAUGGCAAUUUGGCAAUAAAUGCGGCCAUUAGCGCAUUUUAGGUGUUGCUACCGUACUUCUC .............(((.((((((((((((((.((((((..(((...(((((((((......)))))))))(((.....))))))..)))))).))))))....)))))))))))...... ( -32.80) >DroPer_CAF1 81072 109 + 1 C-----------UUGUUGGCGGUAGUAAAAUUCGUUAAAUGCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUGCCCCCAUUAACUCAGUUUAGGUGUUGCCACCGCACUUUUC .-----------......(((((.((((....(.((((((......(((((((((......)))))))))(((.....))).............)))))))..)))).)))))....... ( -22.30) >consensus U___________UUACUGGCGGCAGUAAAAUUCGUUAAACGCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUGCGGCCAUUAACUCAUUUUAGGUGUUGCCACCGCACUUUUC ................((((((((.((((((..(((((..(((...(((((((((......)))))))))(((.....))))))..)))))..))))).).))))))))........... (-24.58 = -24.25 + -0.33)



| Location | 12,697,287 – 12,697,377 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 95 |
| Reading direction | forward |
| Mean pairwise identity | 84.33 |
| Mean single sequence MFE | -18.02 |
| Consensus MFE | -16.02 |
| Energy contribution | -15.38 |
| Covariance contribution | -0.64 |
| Combinations/Pair | 1.22 |
| Mean z-score | -1.36 |
| Structure conservation index | 0.89 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561534 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12697287 90 + 27905053 GCCCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCUAUUAACUCAUUUUAGGUGUUGCCACCGCACUUUUCUGUUCGUCUC----- (((...(((((((((......)))))))))(((.....))))))...............((((....))))...................----- ( -19.50) >DroVir_CAF1 76806 95 + 1 GCUCAACAAAUUGUUUAUUAUAGCAAUUUGGCAAUAAAUGCGGCUAUUAGCUCAUUUUAGGUGUUGCCACCGCACUUUUCUCAUUUUUUAUCGCC ((....(((((((((......)))))))))((...(((((.(((.....))))))))..((((....))))))...................)). ( -20.90) >DroPse_CAF1 74672 90 + 1 GCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUGCCCCCAUUAACUCAGUUUAGGUGUUGCCACCGCACUUUUCUGUUUAUCCA----- ((..(((((((((((......))))))))((((.....))))............)))..((((....)))))).................----- ( -15.70) >DroWil_CAF1 99158 86 + 1 GCUUAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCCAUUAACUCAUUUUAGGUGUUGCCACCGCACUUUUCCAU----CCA----- ......(((((((((......)))))))))((...(((((.(........).)))))..((((....))))))..........----...----- ( -17.30) >DroMoj_CAF1 86060 94 + 1 GCUAAACAAAUUGUUUAUUAUGGCAAUUUGGCAAUAAAUGCGGCCAUUAGCGCAUUUUAGGUGUUGCUACCGUACUUCUCUCCUCUUUUUUU-CC ......(((((((((......)))))))))........(((((....((((((((.....))).))))))))))..................-.. ( -19.00) >DroPer_CAF1 81101 90 + 1 GCUCAACAAAUUGUUUAUUACAGCAGUUUGGCAAUAAAUGCCCCCAUUAACUCAGUUUAGGUGUUGCCACCGCACUUUUCUGUUUAUCCA----- ((..(((((((((((......))))))))((((.....))))............)))..((((....)))))).................----- ( -15.70) >consensus GCUCAACAAAUUGUUUAUUAUAGCAGUUUGGCAAUAAAUGCGGCCAUUAACUCAUUUUAGGUGUUGCCACCGCACUUUUCUGUUUAUCCA_____ ((....(((((((((......)))))))))(((.....)))..................((((....))))))...................... (-16.02 = -15.38 + -0.64)

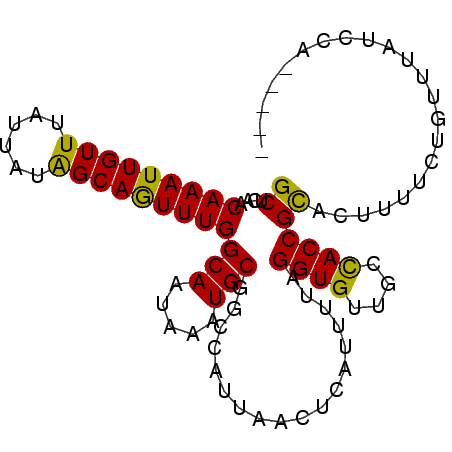

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:40:17 2006