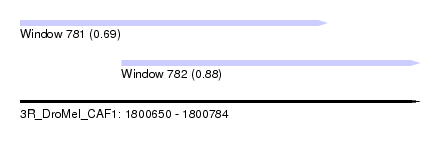
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,800,650 – 1,800,784 |
| Length | 134 |
| Max. P | 0.881959 |

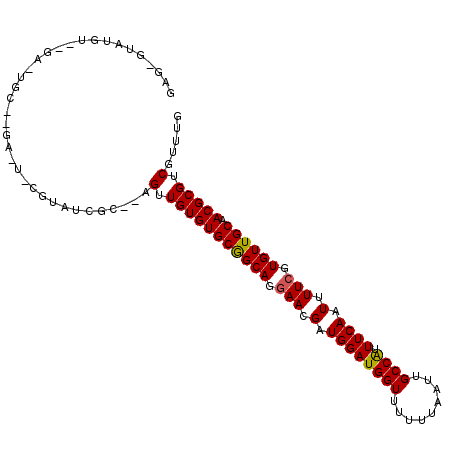
| Location | 1,800,650 – 1,800,753 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 108 |
| Reading direction | forward |
| Mean pairwise identity | 82.05 |
| Mean single sequence MFE | -29.48 |
| Consensus MFE | -19.89 |
| Energy contribution | -19.70 |
| Covariance contribution | -0.19 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.89 |
| Structure conservation index | 0.67 |
| SVM decision value | 0.34 |
| SVM RNA-class probability | 0.693993 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1800650 103 + 27905053 GAG-GUUUGUUCGACUGC--GAGUACCUAUCGC--AGUUGUGUGCGGCAGGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUG ((.-((.....(((((((--((.......))))--)))))..(((((((.(((.(.((((((((.........)))).)))).).))).)))))))..)).))..... ( -34.00) >DroPse_CAF1 99682 92 + 1 GAAUGUAUG--------------UUCGUAUCAC--AGUUGUGUGCGGCAUGAACGAUGGAUGGUUUUUUAAUUGCCGUUUCAAUUUUCGUGUUGCAACGCGUCCUUUG ((((....)--------------))).......--....((((((((((((((.(.((((((((.........)))).)))).).)))))))))).))))........ ( -24.30) >DroEre_CAF1 107368 99 + 1 GAG-GUUUGUGCGACUGC--GA----GUAUCGC--AGUUGUGUGCGGCAGGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUG ((.-((..(..(((((((--(.----....)))--)))))..)((((((.(((.(.((((((((.........)))).)))).).))).))))))...)).))..... ( -35.20) >DroYak_CAF1 110396 97 + 1 GAC------UGCGAGUAC--GA---CGUAUCGCACAGUUGUGUGCGGCAGAAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUCG ...------..(((..((--((---(((..((((((....))))))((((..((((((((((((.........)))).))))....)))).))))...)))))))))) ( -30.90) >DroAna_CAF1 101034 105 + 1 UAG--UAUGUU-GAAUGUUUUUGUACGUAUCGCACAGUUGUGUGCAGCAUGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUG ...--..((((-((((((......)))).))).)))(.(((((((((((((((.(.((((((((.........)))).)))).).)))))))))).))))).)..... ( -28.20) >DroPer_CAF1 102284 92 + 1 GAAUGUAUG--------------UUCGUAUCAC--AGUUGUGUGCGGCAUGAACGAUGGAUGGUUUUUUAAUUGCCGUUUCAAUUUUCGUGUUGCAACGCGUCCUUUG ((((....)--------------))).......--....((((((((((((((.(.((((((((.........)))).)))).).)))))))))).))))........ ( -24.30) >consensus GAG_GUAUGU__GA_UGC__GA_U_CGUAUCGC__AGUUGUGUGCGGCAGGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUG ....................................(.(((((((((((.(((.(.((((((((.........)))).)))).).))).)))))).))))).)..... (-19.89 = -19.70 + -0.19)

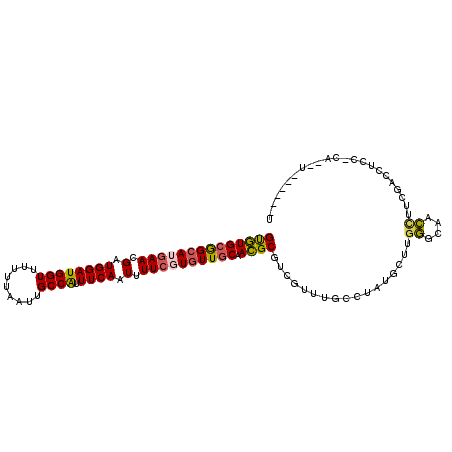
| Location | 1,800,684 – 1,800,784 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 109 |
| Reading direction | forward |
| Mean pairwise identity | 81.12 |
| Mean single sequence MFE | -31.08 |
| Consensus MFE | -22.26 |
| Energy contribution | -22.08 |
| Covariance contribution | -0.18 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.22 |
| Structure conservation index | 0.72 |
| SVM decision value | 0.92 |
| SVM RNA-class probability | 0.881959 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1800684 100 + 27905053 GUGUGCGGCAGGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUGCCUAUGCCUGGGCAACUUUCGACCUCC-CA--U------U ((((((((((.(((.(.((((((((.........)))).)))).).))).)))))).))))((((.(((((((....)))))))....))))....-..--.------. ( -33.70) >DroPse_CAF1 99705 106 + 1 GUGUGCGGCAUGAACGAUGGAUGGUUUUUUAAUUGCCGUUUCAAUUUUCGUGUUGCAACGCGUCCUUUGCCUAGGCUUGCGCAAGCUUCCACCUCG-CA--CUGGAGCU ((((((((((((((.(.((((((((.........)))).)))).).)))))))))).)))).(((..(((..(((...((....)).....))).)-))--..)))... ( -36.40) >DroWil_CAF1 110348 97 + 1 GCAUGCGGCAUGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUUUUACAAUGCCUUUUUUUCUUCUUCUCU------CUUCUUUCCCGACUACU------U ......(((((((((((((((((((.........)))).))))....))))))....))))).................------.................------. ( -15.90) >DroYak_CAF1 110424 100 + 1 GUGUGCGGCAGAAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUCGCCUAUGCCUGGGCAACCUUCGACCUCC-UA--G------C ((((((((((((((....(((((((.........)))))))....)))).)))))).))))((((...(((((....)))))......))))....-..--.------. ( -29.40) >DroAna_CAF1 101070 101 + 1 GUGUGCAGCAUGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUGCCUAUGCUUGGGCAACCUUCGACCCCCACA--U------U ((((((((((((((.(.((((((((.........)))).)))).).)))))))))).))))((((.(((((((....)))))))....)))).......--.------. ( -37.10) >DroPer_CAF1 102307 106 + 1 GUGUGCGGCAUGAACGAUGGAUGGUUUUUUAAUUGCCGUUUCAAUUUUCGUGUUGCAACGCGUCCUUUGCCUAUGCUUGCGCAAGCUUCCACCUCG-CA--CUGGAGCU ((((((((((((((.(.((((((((.........)))).)))).).)))))))))).)))).(((..(((...((...((....))...))....)-))--..)))... ( -34.00) >consensus GUGUGCGGCAUGAACGAUGGAUGGUUUUUUAAUUGCCAUUUCAAUUUUCGUGUUGCAACGCGUCGUUUGCCUAUGCUUGGGCAACCUUCGACCUCC_CA__U______U ((((((((((((((.(.((((((((.........)))).)))).).)))))))))).)))).................((....))....................... (-22.26 = -22.08 + -0.18)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:19 2006