
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
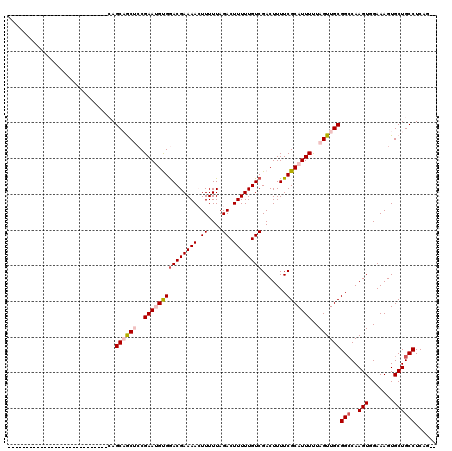
| Location | 12,622,779 – 12,622,869 |
| Length | 90 |
| Max. P | 0.719272 |

| Location | 12,622,779 – 12,622,869 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -25.62 |
| Consensus MFE | -19.39 |
| Energy contribution | -20.28 |
| Covariance contribution | 0.89 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.33 |
| Structure conservation index | 0.76 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.719272 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
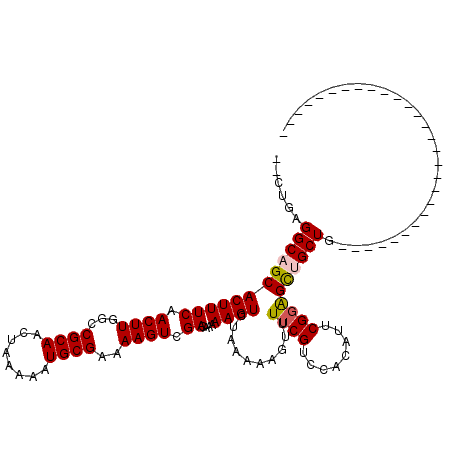
>3R_DroMel_CAF1 12622779 90 + 27905053 ----------------------------CAGCAGCUCCGAAUGUGGACGAAAACUUUUUAGACUUUUUGUCGACUUUUCGCAUUUUUAGUUGCGGCCAAGUGGAAAGUGCUGCCUCAG-- ----------------------------..(((((((((...((.((((((((.((....)).)))))))).))...(((((........))))).....))))....))))).....-- ( -25.00) >DroPse_CAF1 111760 118 + 1 CUCCUCCUCUUCUUUUCUCCUCUCCUUGAAGCCACCCCGAAAGCGGACGAAAAGUUUUUAGACUUUUUGUCGACUUUUCGCAUUUCUAGUUGCGGCCAAGUUGAAAGUGCUGCCUCAG-- ..........................(((.((((((((....).)).(((((((((....)))))))))(((((((.(((((........)))))..)))))))..)))..)).))).-- ( -27.70) >DroSim_CAF1 105951 92 + 1 ----------------------------CAGCAGCUCCGAAUGUGGACGAAAACUUUUUAGACUUUUUGUCGACUUUUCGCAUUUUUAGUUGCGGACAAGUGGAAAGUGCUGCCUCAGCA ----------------------------..(((((((((....)))).....(((((((.(((.....))).((((((((((........)))))).))))))))))))))))....... ( -26.70) >DroYak_CAF1 105512 90 + 1 ----------------------------CAGCAGCUCCGAAUGUGAACGAAAACUUUUUAGACUUUUUGUCGACUUUUCGCAUUUUUAGUUGCGGCCAAGUGGAAAGUGCUGCCUCAG-- ----------------------------..((((((..(((((((((.(((.....))).(((.....))).....)))))))))..))))))(((..(((.......))))))....-- ( -22.90) >DroAna_CAF1 101052 90 + 1 ----------------------------CAGCAGCUACGAAUGUGGACGAAAACUUUUUAGACUUUUUGUCGACUUUUCGCAUUUUUAGUUGCGGCCAAGUUGAAAGUGCUGCCUCAG-- ----------------------------..(((((((.(((((((((((((((.((....)).)))))))).......))))))).)))))))(((..(((.......))))))....-- ( -25.91) >DroPer_CAF1 118038 118 + 1 CUCCUCCUCUUCUUUUCUCCUCUCCUUCAAGCCACCCCGAAAGCGGACGAAAAGUUUUUAGACUUUUUGUCGACUUUUCGCAUUUCUAGUUGCGGCCAAGUUGAAAGUGCUGCCUCAG-- .............................((((.((.(....).)).(((((((((....)))))))))(((((((.(((((........)))))..)))))))..).))).......-- ( -25.50) >consensus ____________________________CAGCAGCUCCGAAUGUGGACGAAAACUUUUUAGACUUUUUGUCGACUUUUCGCAUUUUUAGUUGCGGCCAAGUGGAAAGUGCUGCCUCAG__ ..............................((((((..(((((((((((((((.((....)).)))))))).......)))))))..))))))(((..(((.......))))))...... (-19.39 = -20.28 + 0.89)


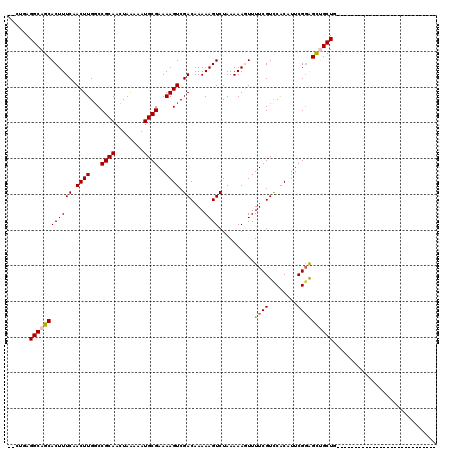
| Location | 12,622,779 – 12,622,869 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | reverse |
| Mean pairwise identity | 79.96 |
| Mean single sequence MFE | -24.12 |
| Consensus MFE | -17.44 |
| Energy contribution | -17.50 |
| Covariance contribution | 0.06 |
| Combinations/Pair | 1.08 |
| Mean z-score | -1.06 |
| Structure conservation index | 0.72 |
| SVM decision value | -0.04 |
| SVM RNA-class probability | 0.510880 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12622779 90 - 27905053 --CUGAGGCAGCACUUUCCACUUGGCCGCAACUAAAAAUGCGAAAAGUCGACAAAAAGUCUAAAAAGUUUUCGUCCACAUUCGGAGCUGCUG---------------------------- --....(((((((((((..((((...((((........))))..)))).(((.....)))...))))).....(((......))))))))).---------------------------- ( -22.30) >DroPse_CAF1 111760 118 - 1 --CUGAGGCAGCACUUUCAACUUGGCCGCAACUAGAAAUGCGAAAAGUCGACAAAAAGUCUAAAAACUUUUCGUCCGCUUUCGGGGUGGCUUCAAGGAGAGGAGAAAAGAAGAGGAGGAG --...........(((((.....((((((..........(((((((((.(((.....))).....)))))))))(((....))).)))))).....)))))................... ( -28.50) >DroSim_CAF1 105951 92 - 1 UGCUGAGGCAGCACUUUCCACUUGUCCGCAACUAAAAAUGCGAAAAGUCGACAAAAAGUCUAAAAAGUUUUCGUCCACAUUCGGAGCUGCUG---------------------------- ......(((((((((((..((((...((((........))))..)))).(((.....)))...))))).....(((......))))))))).---------------------------- ( -22.30) >DroYak_CAF1 105512 90 - 1 --CUGAGGCAGCACUUUCCACUUGGCCGCAACUAAAAAUGCGAAAAGUCGACAAAAAGUCUAAAAAGUUUUCGUUCACAUUCGGAGCUGCUG---------------------------- --....(((((((((((..((((...((((........))))..)))).(((.....)))...))))).((((........)))))))))).---------------------------- ( -21.60) >DroAna_CAF1 101052 90 - 1 --CUGAGGCAGCACUUUCAACUUGGCCGCAACUAAAAAUGCGAAAAGUCGACAAAAAGUCUAAAAAGUUUUCGUCCACAUUCGUAGCUGCUG---------------------------- --....((((((.........((((......))))...((((((..((.(((.((((.(......).)))).))).)).)))))))))))).---------------------------- ( -20.10) >DroPer_CAF1 118038 118 - 1 --CUGAGGCAGCACUUUCAACUUGGCCGCAACUAGAAAUGCGAAAAGUCGACAAAAAGUCUAAAAACUUUUCGUCCGCUUUCGGGGUGGCUUGAAGGAGAGGAGAAAAGAAGAGGAGGAG --...........(((((.((((...((((........))))..)))).(((.....)))......((((((.(((.((((((((....))))))))...)))))))))....))))).. ( -29.90) >consensus __CUGAGGCAGCACUUUCAACUUGGCCGCAACUAAAAAUGCGAAAAGUCGACAAAAAGUCUAAAAAGUUUUCGUCCACAUUCGGAGCUGCUG____________________________ ......((((((((((((.((((...((((........))))..)))).))....))))..........((((........))))))))))............................. (-17.44 = -17.50 + 0.06)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:51 2006