
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,614,850 – 12,615,004 |
| Length | 154 |
| Max. P | 0.991613 |

| Location | 12,614,850 – 12,614,964 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | forward |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -50.28 |
| Consensus MFE | -34.29 |
| Energy contribution | -34.97 |
| Covariance contribution | 0.67 |
| Combinations/Pair | 1.33 |
| Mean z-score | -3.60 |
| Structure conservation index | 0.68 |
| SVM decision value | 2.28 |
| SVM RNA-class probability | 0.991613 |
| Prediction | RNA |
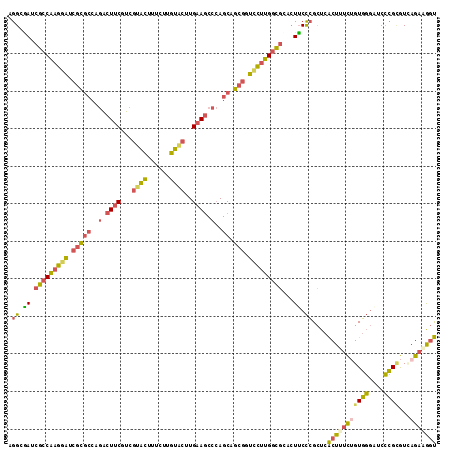
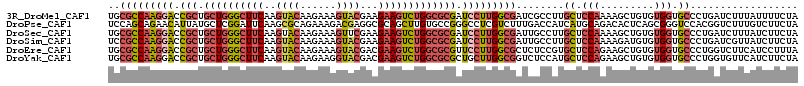
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12614850 114 + 27905053 ACUUUCUAACACGGGAUCCCACAGAAAGUGAGCGGGAAGUGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGAAGAAGUCUGGCGCGAUCCUUGGCGAUCGCCU (((((((.....((....))..)))))))..((((......(((((((((.(((.((..((((((..((((......))))...))))))..))))).))))))))).)))).. ( -52.90) >DroPse_CAF1 101880 114 + 1 GGCAACUGAUGCUCGAGGGAACGAGAAGCGAACUAGCACUCCAGCAGAACAUUAUGCUCGGAUUCAAGCGCAGAAAGACGAGGCGCAGCUUGUGCCGGGCCUCGUCUUUGACCA (((..(((.((((.(((..........((......))..(((((((........)))).)))))).)))))))(((((((((((.(.((....)).).)))))))))))).)). ( -44.20) >DroSec_CAF1 97307 114 + 1 ACCUUCUCACACGGGAUCCCACAGAAAGUGAGCGGGAAGUGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUUCGAAGAAGUCUGGCGCGAUCCUUGGCGAUUGCCU .(((.(((((..((....)).......))))).))).....(((((((((.(((.((..((((((..(.((......)).)...))))))..))))).)))))))))....... ( -47.90) >DroSim_CAF1 97873 114 + 1 ACCUUCUCACGCGGGAACCCACAGAAAGUGAGCGGGAAGUCCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGAAGAAGUCUGGCGCGAUCCUUGGCGAUUGCCU .(((.((((((.((....)).).....))))).))).....(((((((((.(((.((..((((((..((((......))))...))))))..))))).)))))))))....... ( -53.90) >DroEre_CAF1 96752 114 + 1 ACCAUCUGACGCGGGAACCCACUGAAAGUGAGCGGGAAGUGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGACGAAGUCUGGCGCGUUCCUUGGCGCUCUCCG .........(((((....))((.....))..)))(((.(.((((((((((.(((.((..((((((..((((......))))...))))))..))))).)))))))))).)))). ( -56.50) >DroYak_CAF1 97187 114 + 1 ACCAUCUGACCCGGGAUCCCACAGACAGUGAGCGGGAAGUGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAGGUACGACGAAGUCUGGCGCGCUGCUUGGCGGUCUCCA .......((((.....((((.(.........).)))).....((((((.(.(((.((..((((((..((((......))))...))))))..))))).).)))))))))).... ( -46.30) >consensus ACCAUCUGACGCGGGAUCCCACAGAAAGUGAGCGGGAAGUGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGAAGAAGUCUGGCGCGAUCCUUGGCGAUCGCCU ............((....))..............((.....(((((((((.(((.((((((((((..((((......))))...))))))))))))).)))))))))....)). (-34.29 = -34.97 + 0.67)


| Location | 12,614,850 – 12,614,964 |
|---|---|
| Length | 114 |
| Sequences | 6 |
| Columns | 114 |
| Reading direction | reverse |
| Mean pairwise identity | 77.08 |
| Mean single sequence MFE | -45.68 |
| Consensus MFE | -30.42 |
| Energy contribution | -30.18 |
| Covariance contribution | -0.24 |
| Combinations/Pair | 1.50 |
| Mean z-score | -2.50 |
| Structure conservation index | 0.67 |
| SVM decision value | 1.96 |
| SVM RNA-class probability | 0.984090 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12614850 114 - 27905053 AGGCGAUCGCCAAGGAUCGCGCCAGACUUCUUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCACUUCCCGCUCACUUUCUGUGGGAUCCCGUGUUAGAAAGU .((.((.(((((((((((((((..(.((((...((((......))))..)))).)..)).)))))))))))))....))))....(((((((((((....)))...)))))))) ( -48.20) >DroPse_CAF1 101880 114 - 1 UGGUCAAAGACGAGGCCCGGCACAAGCUGCGCCUCGUCUUUCUGCGCUUGAAUCCGAGCAUAAUGUUCUGCUGGAGUGCUAGUUCGCUUCUCGUUCCCUCGAGCAUCAGUUGCC .(((.(((((((((((.((((....)))).)))))))))))(((.((((((....((((.....))))....((((((......))))))........))))))..)))..))) ( -42.70) >DroSec_CAF1 97307 114 - 1 AGGCAAUCGCCAAGGAUCGCGCCAGACUUCUUCGAACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCACUUCCCGCUCACUUUCUGUGGGAUCCCGUGUGAGAAGGU ..((..((((((((((((((((..(.((((...(.((......)).)..)))).)..)).))))))))))))((((.((((((.........))))))....))))..))..)) ( -43.80) >DroSim_CAF1 97873 114 - 1 AGGCAAUCGCCAAGGAUCGCGCCAGACUUCUUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGGACUUCCCGCUCACUUUCUGUGGGUUCCCGCGUGAGAAGGU ......((((((((((((((((..(.((((...((((......))))..)))).)..)).))))))))))))))....((..(((((.....((((....)))))))))..)). ( -49.90) >DroEre_CAF1 96752 114 - 1 CGGAGAGCGCCAAGGAACGCGCCAGACUUCGUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCACUUCCCGCUCACUUUCAGUGGGUUCCCGCGUCAGAUGGU (((.((((((((((((.(((((..(.(((((..((((......)))).))))).)..)).))).))))))))))....((((((.......)))))).)))))........... ( -48.40) >DroYak_CAF1 97187 114 - 1 UGGAGACCGCCAAGCAGCGCGCCAGACUUCGUCGUACCUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCACUUCCCGCUCACUGUCUGUGGGAUCCCGGGUCAGAUGGU ....((((((((((.(.(((((..(.(((((..((((......)))).))))).)..)).))).).)))))).....((((((.........)))))).....))))....... ( -41.10) >consensus AGGCGAUCGCCAAGGAUCGCGCCAGACUUCGUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCACUUCCCGCUCACUUUCUGUGGGAUCCCGCGUCAGAAGGU .((.((.(((((((((.(((((..(.((((...((((......))))..)))).)..)).))).)))))))))...)).))....(((((((((((....))))...))))))) (-30.42 = -30.18 + -0.24)


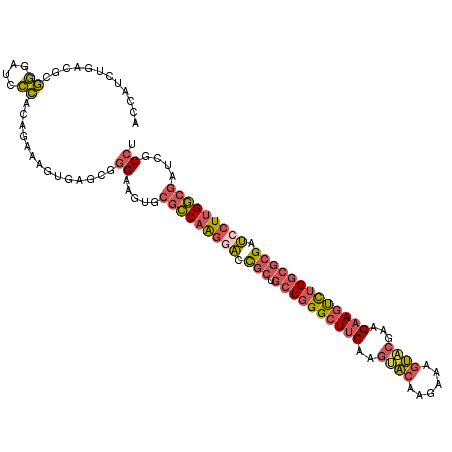
| Location | 12,614,889 – 12,615,004 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | forward |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -45.58 |
| Consensus MFE | -32.65 |
| Energy contribution | -33.93 |
| Covariance contribution | 1.28 |
| Combinations/Pair | 1.29 |
| Mean z-score | -2.57 |
| Structure conservation index | 0.72 |
| SVM decision value | 1.68 |
| SVM RNA-class probability | 0.971871 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12614889 115 + 27905053 UGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGAAGAAGUCUGGCGCGAUCCUUGGCGAUCGCCUUGCUCCAAAAGCUGUGUGGUGCCCUGAUCUUUAUUUUCUA ...((((((((.(((.((..((((((..((((......))))...))))))..))))).))))))))(((((....((.(((.........))).))..)))))........... ( -48.50) >DroPse_CAF1 101919 115 + 1 UCCAGCAGAACAUUAUGCUCGGAUUCAAGCGCAGAAAGACGAGGCGCAGCUUGUGCCGGGCCUCGUCUUUGACCAUCAUGCAGACACUCAGCGGGUCCACGGUCUUUGUCUUCUA (((((((........)))).)))...(((.(((((((((((((((.(.((....)).).)))))))))))(((((((.(((.(.....).))))))....)))).)))))))... ( -39.10) >DroSec_CAF1 97346 115 + 1 UGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUUCGAAGAAGUCUGGCGCGAUCCUUGGCGAUUGCCUUGCUCCAAAAGCUGUGUGGUGCCCUGAUCUUUAUCUUCUA ..(((((((((.(((.((..((((((..(.((......)).)...))))))..))))).)))))))))((..(...(((.....)))...)..)).................... ( -42.20) >DroSim_CAF1 97912 115 + 1 UCCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGAAGAAGUCUGGCGCGAUCCUUGGCGAUUGCCUUGCUCCAAAAGAUGUGUGGUGCCCUGAUCGUUAUCUUCUA ...((((((((.(((.((..((((((..((((......))))...))))))..))))).))))))))(((((....((.(((.........))).))..)))))........... ( -46.20) >DroEre_CAF1 96791 115 + 1 UGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGACGAAGUCUGGCGCGUUCCUUGGCGCUCUCCGUGCUCCAGAAGCUGUGUGGUGCCCUGGUCUUCAUCCUUUA .((((((((((.(((.((..((((((..((((......))))...))))))..))))).))))))))))...(((.((.(((.(.....).))).))..)))............. ( -52.00) >DroYak_CAF1 97226 115 + 1 UGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAGGUACGACGAAGUCUGGCGCGCUGCUUGGCGGUCUCCAUGCUCCAGAAGCUGUGUGGUGCCCUGGUGUUCAUCUUCUA .((((((.(((((((.((..((((((..((((......))))...))))))..))(((((..((((.(((......))))))).))).))))))).)).)))))).......... ( -45.50) >consensus UGCGCCAAGGACCGCUGCUGGGCUUCAAGUACAAGAAAGUACGAAGAAGUCUGGCGCGAUCCUUGGCGAUCGCCUUGCUCCAAAAGCUGUGUGGUGCCCUGAUCUUUAUCUUCUA ..(((((((((.(((.((((((((((..((((......))))...))))))))))))).)))))))))........((.(((.........))).)).................. (-32.65 = -33.93 + 1.28)


| Location | 12,614,889 – 12,615,004 |
|---|---|
| Length | 115 |
| Sequences | 6 |
| Columns | 115 |
| Reading direction | reverse |
| Mean pairwise identity | 78.72 |
| Mean single sequence MFE | -41.28 |
| Consensus MFE | -25.73 |
| Energy contribution | -25.85 |
| Covariance contribution | 0.12 |
| Combinations/Pair | 1.40 |
| Mean z-score | -1.99 |
| Structure conservation index | 0.62 |
| SVM decision value | 0.95 |
| SVM RNA-class probability | 0.888175 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12614889 115 - 27905053 UAGAAAAUAAAGAUCAGGGCACCACACAGCUUUUGGAGCAAGGCGAUCGCCAAGGAUCGCGCCAGACUUCUUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCA ...........((((.((...)).....(((((......)))))))))((((((((((((((..(.((((...((((......))))..)))).)..)).))))))))))))... ( -43.10) >DroPse_CAF1 101919 115 - 1 UAGAAGACAAAGACCGUGGACCCGCUGAGUGUCUGCAUGAUGGUCAAAGACGAGGCCCGGCACAAGCUGCGCCUCGUCUUUCUGCGCUUGAAUCCGAGCAUAAUGUUCUGCUGGA (((.(((((..(((((..(((.(.....).)))..).....))))(((((((((((.((((....)))).)))))))))))....(((((....)))))....)).))).))).. ( -44.20) >DroSec_CAF1 97346 115 - 1 UAGAAGAUAAAGAUCAGGGCACCACACAGCUUUUGGAGCAAGGCAAUCGCCAAGGAUCGCGCCAGACUUCUUCGAACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCA .....(((....)))...((........(((((......)))))....((((((((((((((..(.((((...(.((......)).)..)))).)..)).)))))))))))))). ( -36.70) >DroSim_CAF1 97912 115 - 1 UAGAAGAUAACGAUCAGGGCACCACACAUCUUUUGGAGCAAGGCAAUCGCCAAGGAUCGCGCCAGACUUCUUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGGA .....(((....)))...((.(((.........))).)).......((((((((((((((((..(.((((...((((......))))..)))).)..)).)))))))))))))). ( -41.60) >DroEre_CAF1 96791 115 - 1 UAAAGGAUGAAGACCAGGGCACCACACAGCUUCUGGAGCACGGAGAGCGCCAAGGAACGCGCCAGACUUCGUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCA .............(((((((........)).)))))..........((((((((((.(((((..(.(((((..((((......)))).))))).)..)).))).)))))))))). ( -45.20) >DroYak_CAF1 97226 115 - 1 UAGAAGAUGAACACCAGGGCACCACACAGCUUCUGGAGCAUGGAGACCGCCAAGCAGCGCGCCAGACUUCGUCGUACCUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCA .............(((((((........)).))))).((..(....).((((((.(.(((((..(.(((((..((((......)))).))))).)..)).))).).)))))))). ( -36.90) >consensus UAGAAGAUAAAGACCAGGGCACCACACAGCUUCUGGAGCAAGGCGAUCGCCAAGGAUCGCGCCAGACUUCGUCGUACUUUCUUGUACUUGAAGCCCAGCAGCGGUCCUUGGCGCA .............((...((.(((.........))).))..))....(((((((((.(((((..(.((((...((((......))))..)))).)..)).))).))))))))).. (-25.73 = -25.85 + 0.12)



Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:48 2006