| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,604,525 – 12,604,630 |
| Length | 105 |
| Max. P | 0.720894 |

| Location | 12,604,525 – 12,604,630 |
|---|---|
| Length | 105 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 75.58 |
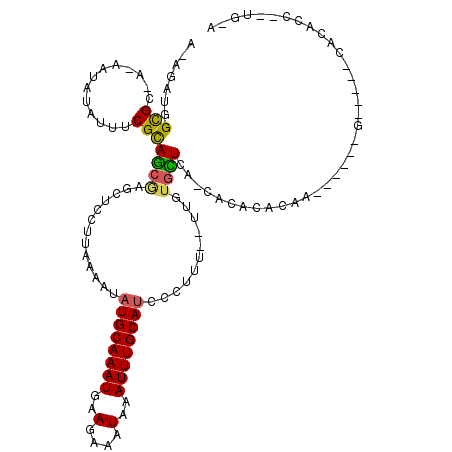
| Mean single sequence MFE | -22.21 |
| Consensus MFE | -7.80 |
| Energy contribution | -7.22 |
| Covariance contribution | -0.58 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.33 |
| Structure conservation index | 0.35 |
| SVM decision value | 0.40 |
| SVM RNA-class probability | 0.720894 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
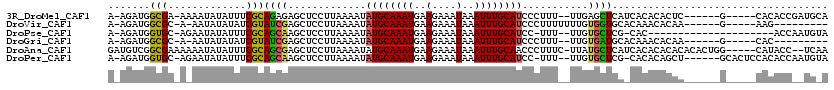
>3R_DroMel_CAF1 12604525 105 + 27905053 A-AGAUGGCGA-AAAAUAUAUUUCGCAGAGAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAUCCCUUU--UUGAGCUCAUCACACACUC------G-----CACACCGAUGCA .-.((((((((-((......)))))).((((((((........((((((((..(....)..))))))))......--..)))))).))...)).))------(-----((......))). ( -28.39) >DroVir_CAF1 95109 97 + 1 A-AGAUGGCGC-A-AAUAUAUAUCGUAUCGAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAUCCCUUUUUUUGUGGUGCACAAACACAA------G-----AAG--------- .-....((.((-.-.((((.....))))...)).)).......((((((((..(....)..)))))))).....((((((((.........)))))------)-----)).--------- ( -17.20) >DroPse_CAF1 90008 93 + 1 A-AGAUGGUGC-AGAAUAUAUUUCGCAGCAAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAUCC-UUU--UUGUGCUCG-CAC---------------------ACCAAUGUA .-...(((((.-.(((.....)))...((.(((....(((((.((((((((..(....)..))))))))..-.))--))).))).)-).)---------------------))))..... ( -22.10) >DroGri_CAF1 90694 95 + 1 A-AGAUGGCGC-A-AAUAUAUAUCGUAUCGAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAUCCCUUU--UUGUGAUGCACAAACACAA------G-----CAC--------- .-.((((.((.-.-.........))))))..(((.........((((((((..(....)..)))))))).....(--(((((...))))))....)------)-----)..--------- ( -14.00) >DroAna_CAF1 83372 112 + 1 GAUGUCGGCGAAAAAAUAUAUUUCGCAGCGAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAACCCUUUC-UUAUGCUCAUCACACACACACACUGG-----CAUACC--UCAA .((((((((((((.......))))))...((((...........(((((((..(....)..)))))))........-....))))...............)))-----)))...--.... ( -23.40) >DroPer_CAF1 95425 108 + 1 A-AGAUGGUGC-AGAAUAUAUUUCGCAGCAAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAUCC-UUU--UUGUGCUCG-CACACAGCU------GCACUCCACACCAAUGUA .-...(((((.-.(((.....)))(((((..............((((((((..(....)..))))))))..-...--.((((....-))))..)))------))......)))))..... ( -28.20) >consensus A_AGAUGGCGC_A_AAUAUAUUUCGCAGCGAGCUCCUUAAAAUAUGCAAAUGAAGAAAUAAAUUUGCAUCCCUUU__UUGUGCUCA_CACACACAA______G_____CACACC__UG_A .......(((.............)))((((.............((((((((..........))))))))...........)))).................................... ( -7.80 = -7.22 + -0.58)
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:41 2006