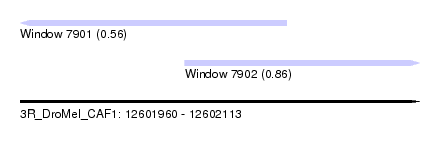
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,601,960 – 12,602,113 |
| Length | 153 |
| Max. P | 0.863014 |

| Location | 12,601,960 – 12,602,062 |
|---|---|
| Length | 102 |
| Sequences | 6 |
| Columns | 106 |
| Reading direction | reverse |
| Mean pairwise identity | 80.26 |
| Mean single sequence MFE | -28.95 |
| Consensus MFE | -21.43 |
| Energy contribution | -24.10 |
| Covariance contribution | 2.67 |
| Combinations/Pair | 1.16 |
| Mean z-score | -1.13 |
| Structure conservation index | 0.74 |
| SVM decision value | 0.06 |
| SVM RNA-class probability | 0.561979 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
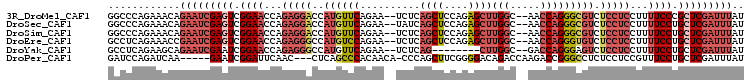
>3R_DroMel_CAF1 12601960 102 - 27905053 GGCCCAGAAACAGAAUCGAGUCGGAACCAGAGGACCAUGUUCAGAA--UCUCAGCUCCAGAGCUUGGC--AACCAGGGCGUCUCCUCCUUUUCCCGCUCGAUUUAU ............(((((((((.((((...(((((..((((((....--....((((....))))(((.--..))))))))).)))))...)))).))))))))).. ( -32.50) >DroSec_CAF1 84534 102 - 1 GGCCCAGAAACAGAAUCGAGUCGGAACCAGAGGACCAUGUUCAGAA--UAUCAGCUCCAGAGCUUGGC--AACCAGGGCGUCUCCUCCUUUUCCUGCUCGAUUUAU ............(((((((((.((((...(((((..((((((....--....((((....))))(((.--..))))))))).)))))...)))).))))))))).. ( -32.50) >DroSim_CAF1 84829 102 - 1 GGCCCAGAAACAGAAUCGAGUCGGAACCAGAGGACCAUGUUCAGAA--UCUCAGCUCCAGAGCUUGGC--AACCAGGGCGUCUCCUCCUUUUCCUGCUCGAUUUAU ............(((((((((.((((...(((((..((((((....--....((((....))))(((.--..))))))))).)))))...)))).))))))))).. ( -32.50) >DroEre_CAF1 83093 102 - 1 GCCUCAGAAACCGAAUCGAGUCGGAACCAGAGGGCCAUGUCCAGAA--UCUCAGCUCCAGAGCUUGGC--AACCAGGGUGUCUCCUCCUUUUCCUGCUCGAUUUAU ............(((((((((.((((...(((((..((..((....--....((((....))))(((.--..)))))..)).)))))...)))).))))))))).. ( -31.20) >DroYak_CAF1 83754 94 - 1 GCCUCAGAAGCAGAAUCGAAUCGGAACCAGAGGGCCAUGUUCAGAA--UCUCAG--------CUUGGC--GACCAGGGAGUCUCCUCCUUUUCCUGCUCGAUUUAU ((.......)).(((((((...((((...((((((((.(((.((..--.)).))--------).))))--(((......))).))))...))))...))))))).. ( -25.90) >DroPer_CAF1 93204 97 - 1 GAUCCAGAUCAA-----GAAUCGGAUUCAAC---CUCAGCCCACAACA-CCCAGCUUCGGGGACAGACCAAGACCGGGCCUCUCCUCCGUUUCCUGCUCGAUUUAU ......((.((.-----(((.((((.....(---(..(((........-....)))..))(((.((.((......))..)).))))))).))).)).))....... ( -19.10) >consensus GGCCCAGAAACAGAAUCGAGUCGGAACCAGAGGACCAUGUUCAGAA__UCUCAGCUCCAGAGCUUGGC__AACCAGGGCGUCUCCUCCUUUUCCUGCUCGAUUUAU ............(((((((((.((((...(((((..((((((..........((((....))))(((.....))))))))).)))))...)))).))))))))).. (-21.43 = -24.10 + 2.67)

| Location | 12,602,023 – 12,602,113 |
|---|---|
| Length | 90 |
| Sequences | 6 |
| Columns | 111 |
| Reading direction | forward |
| Mean pairwise identity | 73.16 |
| Mean single sequence MFE | -37.37 |
| Consensus MFE | -21.28 |
| Energy contribution | -23.28 |
| Covariance contribution | 2.00 |
| Combinations/Pair | 1.23 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.57 |
| SVM decision value | 0.84 |
| SVM RNA-class probability | 0.863014 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
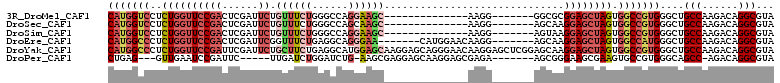
>3R_DroMel_CAF1 12602023 90 + 27905053 CAUGGUCCUCUGGUUCCGACUCGAUUCUGUUUCUGGGCCAGGAAGC--------------AAGG-------GGCGCGGAGCUAGUGGCCGUGGGCUGCCAAGACAGGCGUA (((((.((.(((((((((.(((.....(((((((......))))))--------------).))-------)...))))))))).)))))))....(((......)))... ( -39.10) >DroSec_CAF1 84597 90 + 1 CAUGGUCCUCUGGUUCCGACUCGAUUCUGUUUCUGGGCCAGCAAGC--------------AAGG-------AGCAAGGAGCUAGUGGCCGUGGGCUGCCAAGACAGGCGUA (((((.((.((((((((((......))(((((((..((......))--------------.)))-------)))).)))))))).)))))))....(((......)))... ( -35.50) >DroSim_CAF1 84892 90 + 1 CAUGGUCCUCUGGUUCCGACUCGAUUCUGUUUCUGGGCCAGGAAGC--------------AAGG-------AGUAAGGAGCUAGUGGCCGUGGGCUGCCAAGACAGGCGUA (((((.((.((((((((.((((.....(((((((......))))))--------------)..)-------)))..)))))))).)))))))....(((......)))... ( -37.00) >DroEre_CAF1 83156 97 + 1 CAUGGCCCUCUGGUUCCGACUCGAUUCGGUUUCUGAGGCAGGGAA-------CAUGGAACAAGG-------AGCAAGGAGCUAGUGGCCAUGGGCUGCCAAGACAGGCGUA (((((((..((((((((..((((.((((((((((......)))))-------).)))).)...)-------))...)))))))).)))))))....(((......)))... ( -38.80) >DroYak_CAF1 83809 111 + 1 CAUGGCCCUCUGGUUCCGAUUCGAUUCUGCUUCUGAGGCAUGGAGCAAGGAGCAGGGAACAAGGAGCUCGGAGCAAGGAGCUAGUGGCCGUGGGCUGCCAAGACAGGCGUA (((((((..((((((((..(((((((((((((((...((.....)).)))))).(....)..)))).)))))....)))))))).)))))))....(((......)))... ( -48.60) >DroPer_CAF1 93270 94 + 1 CUGAG---GUUGAAUCCGAUUC-----UUGAUCUGGAUCUG-AAGCGAGGAGCAAGGAGCGAGA-------AGCGGGAAGCGAAGUGCCGUGGGCAGCC-AGACAGGCGUA (((.(---((((..((((.(((-----(((.(((...(((.-......)))....))).)))))-------).))))..(((......)))...)))))-...)))..... ( -25.20) >consensus CAUGGUCCUCUGGUUCCGACUCGAUUCUGUUUCUGGGCCAGGAAGC______________AAGG_______AGCAAGGAGCUAGUGGCCGUGGGCUGCCAAGACAGGCGUA (((((((..((((((((((......)).((((((......))))))..............................)))))))).)))))))....(((......)))... (-21.28 = -23.28 + 2.00)
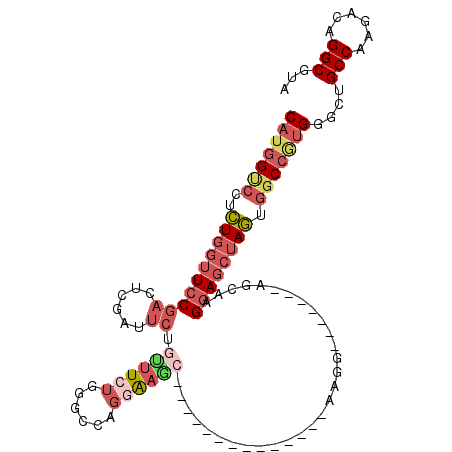
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:40 2006