
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,596,795 – 12,596,924 |
| Length | 129 |
| Max. P | 0.843245 |

| Location | 12,596,795 – 12,596,898 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | forward |
| Mean pairwise identity | 81.74 |
| Mean single sequence MFE | -31.07 |
| Consensus MFE | -18.53 |
| Energy contribution | -19.07 |
| Covariance contribution | 0.53 |
| Combinations/Pair | 1.26 |
| Mean z-score | -2.73 |
| Structure conservation index | 0.60 |
| SVM decision value | 0.76 |
| SVM RNA-class probability | 0.843245 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12596795 103 + 27905053 ------AGAGGA-------GGGCUUCGAGCAAAUCGACG---CUUUGUUUUGUUUACGGCAUUUGUCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAU ------.((((.-------...))))..((((.(((..(---(...((((((((((((((((((..(.....)...)))))))))))))))))).))..))).))))............ ( -30.40) >DroVir_CAF1 86610 110 + 1 ------AGACGCCGGCUCGUGGCAUCAAACAAAUCGGUGG--CGGCGUUUUGUUUACGGCUCUU-UCGAGCUGUAAAAAUGUCGUAAACGCAAUUGUUAUGAAUUGCGGUAAAUUAAAU ------...(((((..(.((........)).)..)))))(--(((((((((...((((((((..-..))))))))))))))))))...(((((((......)))))))........... ( -38.50) >DroPse_CAF1 82524 110 + 1 CAAGACAGAGAG-------AGCCAUCAAACAAAUCGACGG--CAGUGUUUUGUUUACGGCAUUUGUCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAU ((.((((.....-------.(((.((.........)).))--)...((((((((((((((((((..(.....)...)))))))))))))))))))))).)).................. ( -28.00) >DroGri_CAF1 83034 109 + 1 ------AGACGGCGGCUCGUGGCAUCAAACAAAUCGACGA--CCGCAUUUUGUUUACGACUUU--UUGAGCUGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGUAAAUUAAAU ------..((.((((.((((((((..........((....--.)).(((((((((((((((((--(((.....)))))).)))))))))))))))))))))).)))).))......... ( -31.20) >DroAna_CAF1 76919 110 + 1 ------AGAGGCGGGC---GGGCGUCAAACAAAUCGACGGCAGUGUGUUUUGUUUACGGCAUUUGUCGAGUUGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAU ------.((((((.((---.(.((((.........)))).).)).))))))(((((((((((((............)))))))))))))....((((..(.....)..))))....... ( -30.30) >DroPer_CAF1 87983 110 + 1 CAAGACAGAAAG-------AGCCAUCAAACAAAUCGACGG--CAGUGUUUUGUUUACGGCAUUUGUCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAU ((.((((.....-------.(((.((.........)).))--)...((((((((((((((((((..(.....)...)))))))))))))))))))))).)).................. ( -28.00) >consensus ______AGAGGG_______AGGCAUCAAACAAAUCGACGG__CAGUGUUUUGUUUACGGCAUUUGUCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAU .....................((((((((((.....((......))((((((((((((((((((..(.....)...)))))))))))))))))))))).)))..)))............ (-18.53 = -19.07 + 0.53)



| Location | 12,596,821 – 12,596,924 |
|---|---|
| Length | 103 |
| Sequences | 6 |
| Columns | 120 |
| Reading direction | forward |
| Mean pairwise identity | 84.10 |
| Mean single sequence MFE | -28.13 |
| Consensus MFE | -20.62 |
| Energy contribution | -20.15 |
| Covariance contribution | -0.47 |
| Combinations/Pair | 1.24 |
| Mean z-score | -2.64 |
| Structure conservation index | 0.73 |
| SVM decision value | 0.53 |
| SVM RNA-class probability | 0.771357 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12596821 103 + 27905053 ---CUUUGUUUUGUUUACGGCAUUUGUCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAUAAACAUCAAAUGCCAAACGC-UUCUGU------------- ---....((((((((((((((((((..(.....)...)))))))))))))))))).............((((..................))))......-......------------- ( -23.97) >DroVir_CAF1 86643 113 + 1 G--CGGCGUUUUGUUUACGGCUCUU-UCGAGCUGUAAAAAUGUCGUAAACGCAAUUGUUAUGAAUUGCGGUAAAUUAAAUAAACAUCAAAUGCCAAACGCCUUUUGUGUCUGCCAG---- (--(((((((((...((((((((..-..))))))))))))))))))...(((((((......))))))).....................((.((.((((.....)))).)).)).---- ( -33.00) >DroPse_CAF1 82556 104 + 1 G--CAGUGUUUUGUUUACGGCAUUUGUCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAUAAACAUCAAAUGCCAAACGC-UUCUGC------------- (--((((((((((((((((((((((..(.....)...))))))))))))))))))........)))))((((..................))))....((-....))------------- ( -27.77) >DroGri_CAF1 83067 115 + 1 A--CCGCAUUUUGUUUACGACUUU--UUGAGCUGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGUAAAUUAAAUAAACAUCAAAUGCCAAAUGU-UUUUGUGUCUGUCCGCCCU (--(((((((((((((((((((((--(((.....)))))).)))))))))))))...........)))))).......(((.(((.((((.((.....))-.))))))).)))....... ( -27.40) >DroWil_CAF1 53440 103 + 1 G--UGGCAUUUUGUUUACGGCAUUU-UCAAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAUAAACAUCAAAUGCCAAAUGC-UUAUGU------------- .--((((((((((((((((((((((-(.........))))))))))))))))))((((..(.....)..)))).................))))).....-......------------- ( -27.20) >DroAna_CAF1 76949 106 + 1 GCAGUGUGUUUUGUUUACGGCAUUUGUCGAGUUGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAUAAACAUCAAAUGCCAAACGC-UUCUGC------------- ((((.((((((((((((((((((((............)))))))))))))))................((((..................)))).)))))-..))))------------- ( -29.47) >consensus G__CGGCGUUUUGUUUACGGCAUUU_UCGAGUAGUAAAAAUGUCGUAAACGAAAUUGUUAUGAAUUGCGGCAAAUUAAAUAAACAUCAAAUGCCAAACGC_UUCUGU_____________ .....((((((((((((((((((((..(.....)...)))))))))))))).................((((..................)))))))))).................... (-20.62 = -20.15 + -0.47)

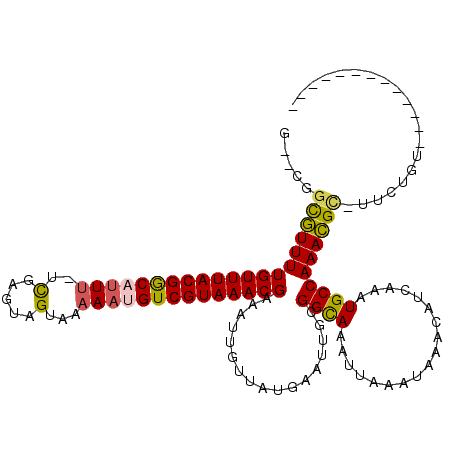

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:37 2006