| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,580,032 – 12,580,138 |
| Length | 106 |
| Max. P | 0.813168 |

| Location | 12,580,032 – 12,580,138 |
|---|---|
| Length | 106 |
| Sequences | 6 |
| Columns | 112 |
| Reading direction | reverse |
| Mean pairwise identity | 80.18 |
| Mean single sequence MFE | -32.53 |
| Consensus MFE | -20.58 |
| Energy contribution | -20.17 |
| Covariance contribution | -0.42 |
| Combinations/Pair | 1.29 |
| Mean z-score | -1.97 |
| Structure conservation index | 0.63 |
| SVM decision value | 0.66 |
| SVM RNA-class probability | 0.813168 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12580032 106 - 27905053 AAUGGCUUUUAUGGCUGACUUCAUGGCUAAUAAAAAUUUGCAUUGUUCGGCCGGGUAGUAUAUUUUCGAUUCUGUUGGUCGAAACCAGAUCGGUUGUG------GCCAUGCC .((((((...(..(((((((...((((((((((.........))))).)))))...)))....((((((((.....))))))))......))))..))------)))))... ( -27.70) >DroPse_CAF1 70188 108 - 1 GAUGGCAUUUAUGGCCGACUUCAUGGCUAAUAAAAAUUAGCAUUGUCUGGCCACAA-AUAUUAUUUC---UCAGUUGGCCGAAAACAGAUCGGCUUUGAGCCGGGCCAGGUC ..((((((((.((((((((......((((((....))))))...))).))))).))-)).......(---((((..((((((.......)))))))))))....)))).... ( -37.00) >DroEre_CAF1 67541 106 - 1 AAUGGCUUUUAUGGCCGACUUCAUGGCUAAUAAAAAUUUGCAUUGUUCGGCCAGGGAGUAUUUUUUCGAUUCUGUUGGUCGAAACCAGAUCGGUUGUG------GCCAUGUC .((((((...(..(((((((((.((((((((((.........))))).))))).)))))....((((((((.....))))))))......))))..))------)))))... ( -33.90) >DroYak_CAF1 67928 106 - 1 AAUGGCUUUUAUGGCCGACUUCAUGGCUAAUAAAAAUUUGCAUUGUUCGGUCGGGGAGUAUUUUUUCGAUUCUGUUGCUCGAAACCAGAUCGGUUGUG------GCCACGUC ..(((((...(..(((((((((.((((((((((.........))))).))))).)))))....((((((.........))))))......))))..))------)))).... ( -26.80) >DroAna_CAF1 66766 104 - 1 AAUGGCUUUAACGGCCGACUUCAUGGCUAAUAAAAAUUUGCAUUGGUUGGCCGCAAAGUAGUUGU--AUUUCAGCUGGCCGAAACCAGAUCGACUUUG------GCCCAGAC ....(((((..(((((((((.....((.(((....))).))...))))))))).)))))(((((.--....)))))(((((((..(.....)..))))------)))..... ( -32.50) >DroPer_CAF1 75535 108 - 1 GAUGGCAUUUAUGGCCGACUUCAUGGCUAAUAAAAAUUAGCAUUGUCUGGCCACAA-AUAUUAUUUC---UCAACUGGCCGAAAACAGAUCGGCUUUGAGCCGGGCCAGGUC ..((((((((.((((((((......((((((....))))))...))).))))).))-)).......(---((((..((((((.......)))))))))))....)))).... ( -37.30) >consensus AAUGGCUUUUAUGGCCGACUUCAUGGCUAAUAAAAAUUUGCAUUGUUCGGCCACAAAGUAUUUUUUCGAUUCAGUUGGCCGAAACCAGAUCGGCUGUG______GCCAGGUC ..((((.....(((((((...((..((.(((....))).))..)).))))))).......................((((((.......)))))).........)))).... (-20.58 = -20.17 + -0.42)

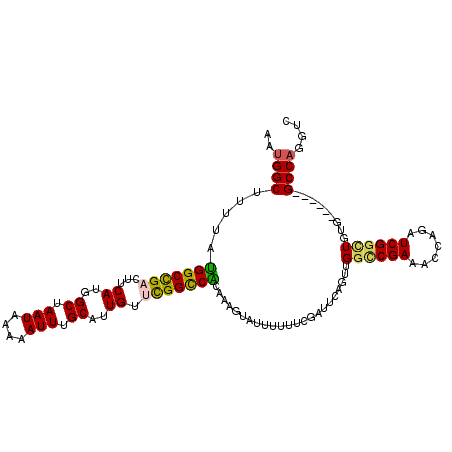
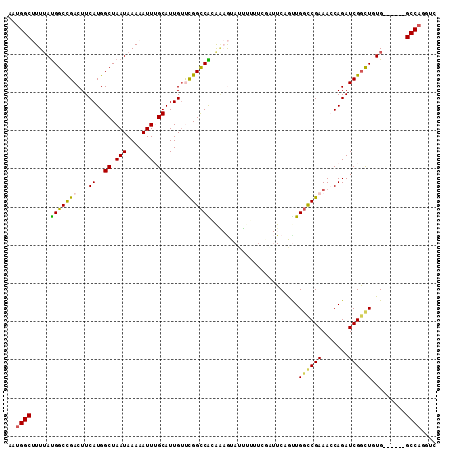
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:32 2006