
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 12,546,851 – 12,546,975 |
| Length | 124 |
| Max. P | 0.740818 |

| Location | 12,546,851 – 12,546,941 |
|---|---|
| Length | 90 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 95.81 |
| Mean single sequence MFE | -19.18 |
| Consensus MFE | -17.38 |
| Energy contribution | -17.26 |
| Covariance contribution | -0.12 |
| Combinations/Pair | 1.07 |
| Mean z-score | -1.27 |
| Structure conservation index | 0.91 |
| SVM decision value | 0.45 |
| SVM RNA-class probability | 0.740818 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12546851 90 + 27905053 ACCGUUCAUUAAGUGUAGCUAAGCA-AACAAACUGCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGGACGUUUUAAUGACA ..((((((((...(((((.......-......)))))..(((((.(((....))).)))))...........))))))))........... ( -18.92) >DroSec_CAF1 35781 91 + 1 ACCGUUCAUUAAGUGUGGCUUAGAAAAACAAACUGCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACA ...((.((((((.((..(.((........)).)..)).....((((((((((.....((((((...)))))))))))))))))))))))). ( -18.40) >DroSim_CAF1 35732 91 + 1 ACCGUUCAUUAAGUGUGGCUUAGAAAAACAAACUGCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGGCA ...(((..(((((.....)))))...))).....((.(((..((((((((((.....((((((...))))))))))))))))..))).)). ( -18.40) >DroEre_CAF1 33591 90 + 1 ACCGUUCAUUAAGUGUGGCUUAGCA-AACAAACUGCAAUUGCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACA ...((.((((((.....((...(((-.......)))....))((((((((((.....((((((...)))))))))))))))))))))))). ( -20.10) >DroYak_CAF1 33073 90 + 1 ACCGCUCAUUAAGUGUGGCUUAGCA-AACAAACUGCAAUUGCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACA .((((.(.....).))))....(((-.......))).((((.((((((((((.....((((((...)))))))))))))))).)))).... ( -20.10) >consensus ACCGUUCAUUAAGUGUGGCUUAGCA_AACAAACUGCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACA ...(.(((((((.(((((..............))))).....((((((((((.....((((((...)))))))))))))))))))))))). (-17.38 = -17.26 + -0.12)



| Location | 12,546,884 – 12,546,975 |
|---|---|
| Length | 91 |
| Sequences | 5 |
| Columns | 91 |
| Reading direction | forward |
| Mean pairwise identity | 97.14 |
| Mean single sequence MFE | -21.00 |
| Consensus MFE | -16.92 |
| Energy contribution | -16.88 |
| Covariance contribution | -0.04 |
| Combinations/Pair | 1.04 |
| Mean z-score | -1.69 |
| Structure conservation index | 0.81 |
| SVM decision value | -0.01 |
| SVM RNA-class probability | 0.526785 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 12546884 91 + 27905053 GCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGGACGUUUUAAUGACACAAUGAUAUGGUAAAUUACGCCAGUGUAAAUGCU (((.......((((((((((.((((((((...)))))))))))))(((.....)))......))))).....(((((....))))).))). ( -19.30) >DroSec_CAF1 35815 91 + 1 GCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACACAAUGAUAUGGUAAAUUACGCCAGUGUAAAUGCU (((..(((((.(((....))).))))).............((((((((.....))).........((((.......)))))))))..))). ( -20.10) >DroSim_CAF1 35766 91 + 1 GCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGGCACAAUGAUAUGGUAAAUUACGCCAGUGUAAAUGCU (((..(((((.(((....))).))))).............((((((((.....))).........((((.......)))))))))..))). ( -19.60) >DroEre_CAF1 33624 91 + 1 GCAAUUGCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACACAAUGAUAUGGUAAAUUACGCCACUGCAAAUGCU (((.((((((((((((((.....((((((...)))))))))))))))).................((((.......))))..)))).))). ( -23.00) >DroYak_CAF1 33106 91 + 1 GCAAUUGCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACACAAUGAUAUGGUAAAUUACGCCACUGCAAAUGCU (((.((((((((((((((.....((((((...)))))))))))))))).................((((.......))))..)))).))). ( -23.00) >consensus GCAAUUCCAACGUGUGUUCACUUUGGAUCAAAGAUUCAAAUGCACGUUUUAAUGACACAAUGAUAUGGUAAAUUACGCCAGUGUAAAUGCU ((((((..((((((((((.....((((((...))))))))))))))))..))).......((...((((.......))))...))..))). (-16.92 = -16.88 + -0.04)

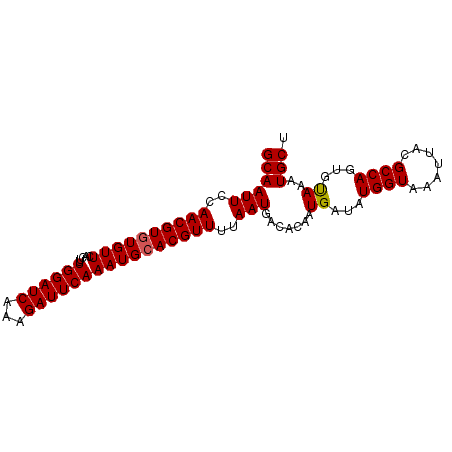
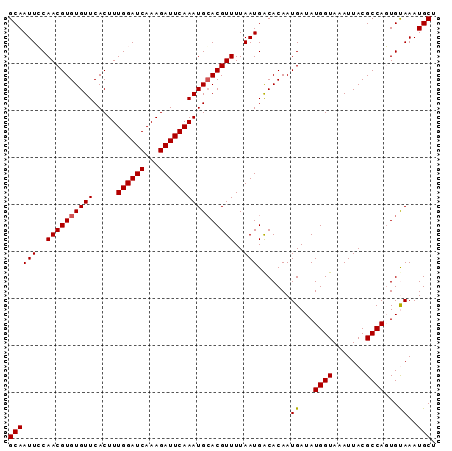
Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 11:39:15 2006