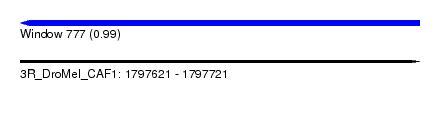
| Sequence ID | 3R_DroMel_CAF1 |
|---|---|
| Location | 1,797,621 – 1,797,721 |
| Length | 100 |
| Max. P | 0.988780 |

| Location | 1,797,621 – 1,797,721 |
|---|---|
| Length | 100 |
| Sequences | 6 |
| Columns | 119 |
| Reading direction | reverse |
| Mean pairwise identity | 76.81 |
| Mean single sequence MFE | -34.04 |
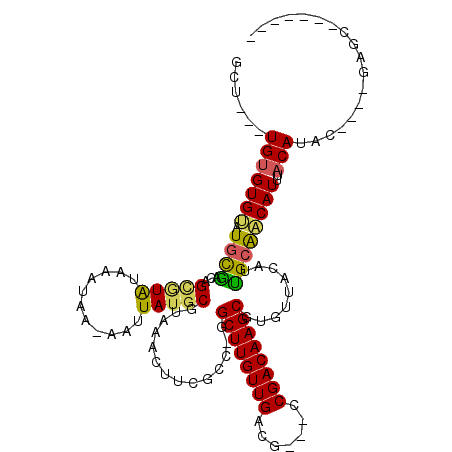
| Consensus MFE | -19.54 |
| Energy contribution | -18.57 |
| Covariance contribution | -0.97 |
| Combinations/Pair | 1.33 |
| Mean z-score | -2.72 |
| Structure conservation index | 0.57 |
| SVM decision value | 2.13 |
| SVM RNA-class probability | 0.988780 |
| Prediction | RNA |
Download alignment: ClustalW | MAF
>3R_DroMel_CAF1 1797621 100 - 27905053 GCU---UGUGUGCCUGCGACCGUGUGUAAAUAA-AAUUAUGCGUAAACUUCGUC-GGCUUGUUGACG---CCGACAAGCGUGUUACAUUGCAGCAUUUACAUGC----GAGC------- (((---((((((.((((((....((((((....-..))))))((((.(((.(((-(((........)---))))))))....)))).))))))......)))))----))))------- ( -31.90) >DroPse_CAF1 97164 109 - 1 GCUCUGUGUGUGUAUGCCACAGCAUAUAAAUAA-AAUUAUGCGUAAAGUUUCCCAGGCUUGUUGACG---CCGACAAGCGUUUUACGUGGCAACAUUCACAUAC----GAGUACGAG-- ((((.(((((((..((((((.(((((.......-...)))))(((((.......(.((((((((...---.)))))))).)))))))))))).....)))))))----)))).....-- ( -40.01) >DroWil_CAF1 108065 104 - 1 GUU---UGUGUGUGUAUGU-AGGUUAUAAAUAACAAUUAUGCGUAAACUUUUCA-GGCUUGUUGAUU---UCGACAAGCGUAUUACAUUGCAACAUUUUCAUACAUAUAAGC------- (((---((((((((((((.-..((((....))))...)))))((..........-.((((((((...---.))))))))(((......))).))........))))))))))------- ( -27.30) >DroYak_CAF1 107436 100 - 1 GCU---UGUGUGCAUGCGACCGUGUGUAAAUAA-AAUUAUGCGUAAACUUCGUC-UGCUUGUUGACG---CCGACAAGCGUGUUACAUUGCGGCAUUUACAUGC----GAGC------- (((---((((((.....(.((((((((((....-......(((.......))).-.((((((((...---.))))))))...))))).)))))).....)))))----))))------- ( -30.20) >DroAna_CAF1 98412 104 - 1 GCC---UGUGUGGCUGUGUGUGCGUGUAAAUAA-AAUUAUGCGUAAACUUCGCCAAGCUUGUUGACGCCGCCGACAAGCGUGUUACAUCGCAACAUUAACAUGC----GAGC------- ((.---((((((...((((.(((((((((....-......(((.......)))...((((((((.......))))))))...))))).))))))))...)))))----).))------- ( -33.10) >DroPer_CAF1 99774 111 - 1 GCUCUGUGUGUGUAUGCCACAGCAUAUAAAUAA-AAUUAUGCGUAAAGUUUCCCAGGCUUGUUGACG---CCGACAAGCGUGUUACGUGGCAACAUUCACAUAC----GAGUACGAGUU ((((.(((((((..((((((.(((((.......-...)))))((((.......((.((((((((...---.)))))))).)))))))))))).....)))))))----))))....... ( -41.71) >consensus GCU___UGUGUGUAUGCGACAGCGUAUAAAUAA_AAUUAUGCGUAAACUUCGCC_GGCUUGUUGACG___CCGACAAGCGUGUUACAUUGCAACAUUUACAUAC____GAGC_______ ......(((((((.((((...(((((...........)))))..............((((((((.......)))))))).........))))))))..))).................. (-19.54 = -18.57 + -0.97)

Generated by rnazCluster.pl (part of RNAz 1.0) on Mon Dec 4 09:44:14 2006